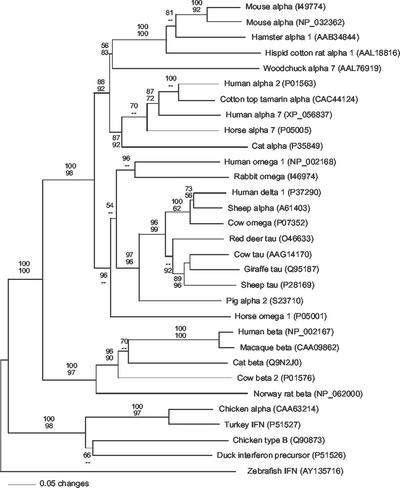
FIG. 6.
Phylogenetic relationships among zfIFN and representative IFN-α, -β, -δ, -ω, and -τ from mammals and birds. Phylogenetic analyses were performed using evolutionary-distance (shown) and maximum-parsimony algorithms. Bootstrap proportions (bp) are presented as the fractional percentage of 100 replicates (top, evolutionary distance; bottom maximum parsimony). Dashes, bp of <50%. Best trees recovered by both algorithms were identical with respect to all well-supported nodes (bp > 70%). The aligned sequence set included 31 sequences of 212 equally weighted positions (185 were parsimony informative). Alignment gaps are treated as missing data. GenBank accession numbers are shown in parentheses.