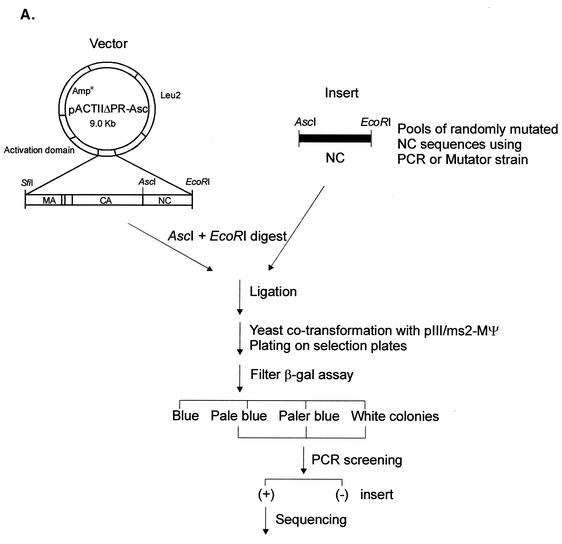
FIG. 4.
(A) Strategy for in vivo screening of random NC mutants using the yeast three-hybrid system. Pools of randomly mutated NC sequences by using either PCR or a mutator strain were swapped for the wt NC sequences of the pACTIIΔPR-Asc protein hybrid plasmid. The ligated DNAs were used to cotransform yeast cells along with the pIII/MS2-MΨ RNA hybrid plasmid and plated onto plates lacking Ura and Leu to select both plasmids. β-Gal activity of each transformant was qualitatively measured by filter β-Gal assays. Colonies with low β-Gal activities were screened for the NC inserts using PCR. The positive (+) clones were sequenced to locate mutations in the NC sequences. (B) Mutational changes resulting from random mutagenesis and in vivo screening. The amino acid sequence of NC is shown at the top, with the numbering starting from the amino terminus of NC. The two CH boxes are underlined, and the Zn-coordinating residues within the CH boxes are shaded. The mutational changes predicted to significantly lower the β-Gal activity are shown in bold type. The asterisk in line 9 represents a mutational change from glutamine to arginine at residue 33 of NC. A stop mutation at Gln69 (line 12) led to the truncation of NC at the C terminus. The relative β-Gal activities indicated are the averages from three independent assays. The mutant with low β-Gal activity (less than 20% of that of ΔPR) are indicated by shading. (C) Additional site-directed mutants. ΔN has residues 1 to 13 of NC deleted; ΔC lacks all of the amino acids following position 68. The G52W mutant which had β-Gal activity of less than 0.2 is shaded.