Figure 3.
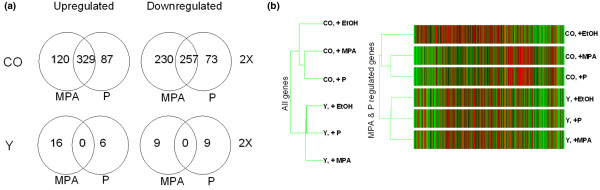
Expression profiles of endogenous PR-regulated genes. Shown are expression profiles of PR-regulated endogenous genes in breast cancer cell lines, with physiological progesterone versus MPA concentrations compared. (a) Venn diagrams showing gene number per condition. PR-positive T47Dco cells (CO) and control PR-negative T47D-Y (Y) cells were treated with 10 nmol/l progesterone or MPA for 6 hours in triplicate, time-separated experiments. Microarray analysis using Affymetrix U-133 2 plus gene chips, displaying about 47,000 genes, was performed. Data were analyzed using Gene Spring software and statistical analysis was conducted using a one-way analysis of variance, with P < 0.05 considered statistically significant for genes regulated at least 2.0-fold. Detailed tables showing complete gene lists are available as Additional files 1 and 2. (b) Dendrograms of PR-regulated genes showing relationships among treatment groups. Gene Spring 6.0 was used to classify all genes 'present' and all MPA- and progesterone-regulated genes. On the left is shown the relatedness among groups for all genes. On the right is a heat map of progestin regulated genes: black = average expression; red = above average; and green = below average. Each column represents a single gene; each row is a cell type and treatment group. DHT, dihydrotestosterone; MPA, medroxyprogesterone acetate; PR, progesterone receptor.
