Abstract
Eukaryotic initiation factor (eIF) 1 maintains the fidelity of initiation codon selection and enables mammalian 43S preinitiation complexes to discriminate against AUG codons with a context that deviates from the optimum sequence GCC(A/G)CC AUGG, in which the purines at −3 and +4 positions are most important. We hypothesize that eIF1 acts by antagonizing conformational changes that occur in ribosomal complexes upon codon–anticodon base-pairing during 48S initiation complex formation, and that the role of −3 and +4 context nucleotides is to stabilize these changes by interacting with components of this complex. Here we report that U and G at +4 both UV-cross-linked to ribosomal protein (rp) S15 in 48S complexes. However, whereas U cross-linked strongly to C1696 and less well to AA1818–1819 in helix 44 of 18S rRNA, G cross-linked exclusively to AA1818–1819. U at −3 cross-linked to rpS5 and eIF2α, whereas G cross-linked only to eIF2α. Results of UV cross-linking experiments and of assays of 48S complex formation done using α-subunit-deficient eIF2 indicate that eIF2α’s interaction with the −3 purine is responsible for recognition of the −3 context position by 43S complexes and suggest that the +4 purine/AA1818–1819 interaction might be responsible for recognizing the +4 position.
Keywords: Ribosome, translation, initiation, nucleotide context, eIF1, eIF2α
Eukaryotic ribosomes locate the initiation codon on most mRNAs by a scanning mechanism. A 43S complex comprising a ribosomal 40S subunit, eukaryotic initiation factors (eIFs) 1, 1A, and 3, and an eIF2-GTP/Met-tRNAMeti complex binds to the 5′-cap-proximal region of mRNA with the help of eIF4A, eIF4B, and eIF4F and scans downstream to the initiation codon to form a 48S complex. Initiation codon recognition and base-pairing with the Met-tRNAMeti anticodon triggers eIF5-mediated hydrolysis of eIF2-bound GTP and, most importantly, subsequent release of phosphate (Algire et al. 2005). The prevailing model is that this leads to release of eIF2-GDP from the 40S subunit, retaining Met-tRNAMeti in the ribosomal P site, after which eIF5B mediates displacement of other factors and joining of the 60S ribosomal subunit to form an 80S ribosome (Pestova et al. 2000; Unbehaun et al. 2004).
The initiation codon is recognized by base-pairing with the anticodon of Met-tRNAMeti (Cigan et al. 1988) and is usually the first AUG triplet from the mRNA’s 5′ end. Scanning 40S subunits can bypass the first AUG triplet if it is <10 nucleotides (nt) from the 5′ end of mRNA or if its context deviates from the optimum sequence GCC(A/G)CCAUGG, particularly at −3 and +4 positions (in bold) (Kozak 1986, 1991). These two context nucleotides are conserved features of mammalian mRNAs and together can enhance translation 20-fold; in yeast, the nucleotide context is less important for initiation codon recognition, and its only common feature is a purine at the −3 position (Kozak 1986; Cavener and Ray 1991). eIF1 enhances the processivity of scanning and plays the key role in ensuring the fidelity of initiation codon selection by enabling 43S complexes to discriminate against 48S complex formation on non-AUG triplets, on AUG triplets located near the 5′ end of mRNA, and on AUG triplets with suboptimal context (Yoon and Donahue 1992; Pestova et al. 1998; Pestova and Kolupaeva 2002). It also ensures the fidelity of initiation codon selection at the later stage of ribosomal subunit joining by inhibiting premature GTP hydrolysis by eIF2 and by coupling initiation codon recognition with activation of eIF2’s GTPase activity (Unbehaun et al. 2004; Valasek et al. 2004; Maag et al. 2005).
eIF1 binds to the interface surface of the 40S subunit between the platform and initiator tRNA, facing the codon–anticodon base pairs but not contacting them directly (Lomakin et al. 2003). This suggests that it promotes scanning and performs its monitoring function indirectly, by influencing the conformation of the platform and the positions of Met-tRNAMeti and mRNA in ribosomal complexes. To explain eIF1’s mechanism of action, we propose that binding of eIF1–43S complexes induces a scanning-competent conformation that is favorable for rejection of codon–anticodon mismatches and does not permit activation of hydrolysis of eIF2-bound GTP by eIF5. However, on recognizing the initiation codon, a 43S complex would have to undergo conformational changes upon base-pairing to form a 48S complex, which would be antagonized by eIF1. In our model, the conformation of an arrested 48S complex would be stabilized by codon–anticodon base-pairing and by elements within mRNA such as 5′-flanking sequences and context nucleotides. If a 48S complex assembled without eIF1 is insufficiently stable, due to the presence of a noncognate initiation codon, poor context or to the absence of 5′-flanking sequences, it dissociates on delayed addition of eIF1 (Pestova and Kolupaeva 2002). An implication of this model is that the role of context nucleotides (particularly of −3 and +4 positions) is to stabilize an arrested ribosomal complex by interacting specifically with its constituents. Hypotheses that context nucleotides interact with 18S rRNA (e.g., Kozak 1986; Cavener and Ray 1991) in a functionally analogous manner to the Shine-Dalgarno interaction of 16S rRNA and prokaryotic mRNAs have not been substantiated.
In this study we used mRNAs with either 4-thiouridine (“thioU”) or 6-thioguanosine (“thioG”) at −3 and +4 positions (hereafter, [−3] and [+4]) for “zero-length” UV cross-linking of ribosomal proteins, 18S rRNA, and initiation factors in 48S complexes to identify nucleotide-specific interactions of purines and pyrimidines at [−3] and [+4] that could account for the nucleotide context rule. U and G at [+4] both cross-linked to ribosomal protein (rp) S15, but whereas U[+4] specifically cross-linked mostly to C1696 and to some extent to AA1818–1819 in helix 44 of 18S rRNA, G[+4] cross-linked exclusively to AA1818–1819. The base specificity of the interaction of the [+4] purine with AA1818–1819 is therefore likely responsible for recognition of this context nucleotide by 43S complexes, so that in addition to monitoring the fidelity of elongator tRNA selection (Ogle et al. 2001), AA1818–1819 might also play a role in initiation codon selection. U[−3] specifically and equally efficiently cross-linked to rpS5 and to eIF2α, whereas G[−3] cross-linked exclusively to eIF2α. The functional involvement of eIF2α in recognizing the [−3] nucleotide was confirmed by assaying 48S complex formation in the presence of α-subunit-deficient eIF2. In the absence of eIF1, the effect of the lack of eIF2α on the efficiency and specificity of 48S complex formation on AUG triplets with different nucleotide contexts was minor. However, in the presence of eIF1, 43S complexes assembled without eIF2α could no longer discriminate the nature of the −3 nucleotide, and 48S complex formation was much less efficient, irrespective of the nucleotide at [−3]. This suggests that interaction of the [−3] nucleotide with eIF2α is generally important for 48S complex formation in the presence of eIF1, but that eIF2α interacts more strongly with a purine than with a pyrimidine residue, increasing the resistance of 48S complexes to dissociation by eIF1, and that this accounts for the −3 nucleotide context rule. The fact that without sucrose density gradient centrifugation 85%–90% of eIF2 remained associated with 48S complexes formed on AUG triplets with G[−3] after eIF5-induced hydrolysis of eIF2-bound GTP could account for the resistance of 48S complexes to eIF1’s dissociating influence after GTP hydrolysis and before the actual ribosomal subunit joining.
Results
48S complex formation on (CAA)nAUG(CAA)m mRNAs containing thioU and thioG
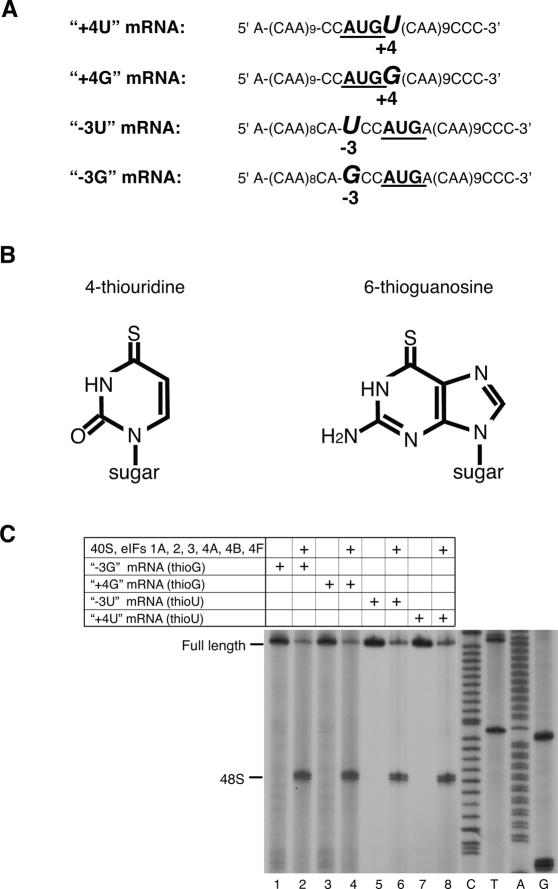
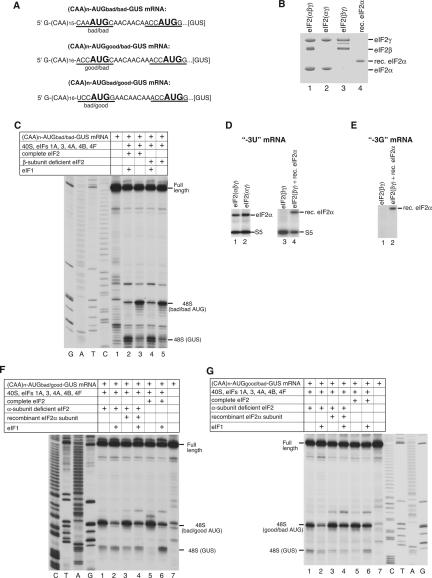
We hypothesized that the role of [−3] and [+4] context nucleotides could be to stabilize conformational changes in 48S complexes that occur upon base-pairing, by interacting with elements of these complexes. We investigated their interactions with components of the 48S complex by UV cross-linking using mRNAs that had a single uridine or guanosine (in addition to the AUG codon) at these positions (Fig. 1A). In two mRNAs the context of AUG triplets was good (purines at [−3] and [+4]) and in two it was suboptimal (a pyrimidine at [−3] or [+4]). Flanking AUG codons with multiple CAA triplets to avoid additional U or G nucleotides also minimized secondary structure and increased initiation efficiency. mRNAs were transcribed in vitro in the presence of thioU or thioG (Fig. 1B), which can be specifically cross-linked to proteins and nucleic acids by low-energy (360-nm) irradiation, yielding “zero-length” cross-links that represent direct contacts with 48S complex constituents. Differences in the specificity/intensity of cross-links between mRNAs containing either thioU or thioG could be indicative of the nucleotide specificity of interactions. ThioG is incorporated less efficiently than thioU into transcripts and may exist in a thiol–thione equilibrium that could lead to its misincorporation (Sergiev et al. 1997; Favre et al. 1998). Toe-printing analysis done in the absence of eIF1 (to avoid potential differences in efficiency of 48S complex formation due to context differences of initiation codons) showed that 48S complex assembled equally and efficiently on mRNAs containing thioG or thioU, which were therefore functional (Fig. 1C).
Figure 1.
48S complex formation on thioU- and thioG-containing mRNAs. (A) Sequences of (CAA)n-AUG-(CAA)m mRNA derivatives containing U or G at −3 and +4 positions (italicized) relative to the A of the initiation codon (bold). (B) Structural formulae of thioU and thioG. (C) Toe-print analysis of 48S complexes assembled as described in Materials and Methods on mRNAs as indicated. Toe-prints due to 48S complexes are shown on the left. Full-length cDNAs are labeled. Lanes C, T, A, G show cDNA sequence corresponding to “+4G” mRNA derived using the same primer as for toe-printing.
Contacts of the nucleotide at position [+4] of mRNA with components of the 48S complex
mRNA transcripts used for UV cross-linking contained thioU or thioG and were labeled with 32P-CTP. 48S complexes assembled from 40S subunits, eIF2, eIF3, eIF4A, eIF4B, eIF4F, eIF1, eIF1A, and Met-tRNAMeti were purified from unincorporated components by sucrose density gradient centrifugation and cross-linked by irradiation at 360 nm. U[+2] and G[+3] of the initiation codon base-pair with the Met-tRNAMeti anticodon and thus cannot cross-link to other components of 48S complexes, so radiolabeling of factors, ribosomal proteins, and 18S rRNA can be attributed exclusively to interactions with [−3] and [+4] nucleotides. In control experiments, thioU[+2] or thioG[+3] did not cross-link to components of 48S complexes assembled on mRNA with U and G only in the initiation codon (V.G. Kolupaeva, A.V. Pisarev, C.U.T. Hellen, and T.V. Pestova, in prep.).
Because eIF1 is a functional analog of prokaryotic IF3 (which causes rearrangement of mRNA on 30S subunits) (La Teana et al. 1995; Shapkina et al. 2000), mRNA cross-linking was assayed in 48S complexes assembled with and without eIF1. RNase-treated samples were analyzed by SDS-PAGE and two-dimensional (2D) gel electrophoresis to identify cross-linked proteins.
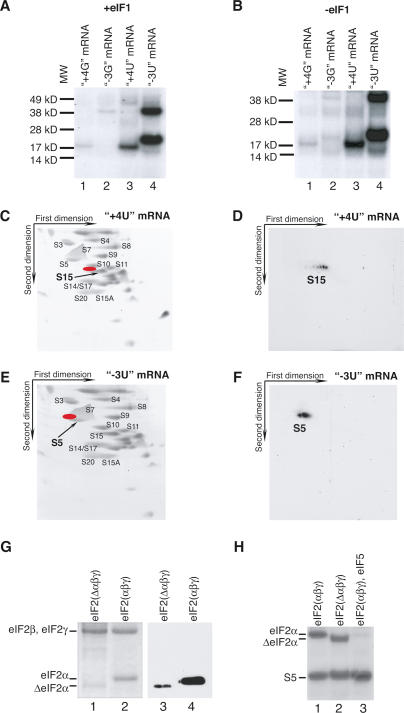
A single protein of the same mobility was cross-linked in 48S complexes assembled with or without eIF1 on mRNAs with thioU or thioG at [+4] (Fig. 2A,B, lanes 1,3). The specificity of UV cross-linking of thioU- and thioG-containing mRNAs did not differ, but consistent with other studies, thioU cross-linked more efficiently than thioG (Nikiforov and Connolly 1992; Fig. 2A,B, lanes 1,3), likely reflecting intrinsic differences in cross-linking efficiencies of these thionucleotides. The low molecular weight of the cross-linked protein indicated that it was a ribosomal protein. Covalently bound mRNA nucleotides cause cross-linked ribosomal proteins to shift “northwest” in 2D gels. Taking this into consideration, cross-linking to [+4] was attributed to rpS15 (Fig. 2C,D). Its identity was confirmed by mass-spectrometry sequencing of EAPPMEKPEVVK and GVDLDQLLDMSYEQLMQLYSAR peptides. rpS15 is a homolog of prokaryotic rpS19, whose position in the crystal structure of the Thermus thermophilus 30S subunit is shown in Figure 4 (see below).
Figure 2.
Contacts of nucleotides at −3 and +4 positions of mRNA with ribosomal proteins and factors in 48S complexes. (A,B) UV cross-linking of 32P-labeled (CAA)n-AUG-(CAA)m mRNAs containing 4-thioU or 6-thioG at [−3] and [+4] as indicated with components of 48S complexes assembled with (A) or without (B) eIF1, assayed by SDS-PAGE and autoradiography. The positions of molecular weight markers (MW) are shown on the left. (C–F) Analysis by 2D electrophoresis of ribosomal proteins UV-cross-linked to 32P-labeled (CAA)n-AUG-(CAA)m mRNAs containing 4-thioU at [+4] (C,D) and at [−3] (E,F). (C,E) Gels of 40S subunit proteins stained with Simply Blue Safe Stain. (D,F) Autoradiographs of gels from C and E. Positions corresponding to radioactive spots (D,F) on stained gels (C,E) are shown in red. The positions of some ribosomal proteins based on sequencing data or according to Madjar et al. (1979) are indicated. (G) eIF2 with full-length (lanes 2,4) and truncated (lanes 1,3) eIF2α assayed by SDS-PAGE and Coomassie staining (lanes 1,2) or immunobloting (lanes 3,4). eIF2 subunits are indicated on the left. (H) UV cross-linking of 32P-labeled (CAA)n-AUG-(CAA)m mRNA derivative containing 4-thioU at [−3] with components of 48S complexes assembled using eIF2 with intact eIF2α (lane 1), eIF2 with truncated eIF2α (lane 2), and eIF2 with intact eIF2α and eIF5 (lane 3), assayed by SDS-PAGE and autoradiography. Positions of eIF2α and rpS5 are indicated on the left.
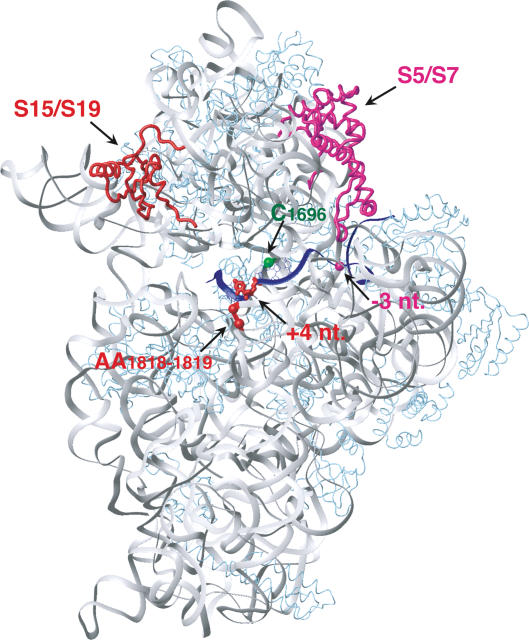
Figure 4.
Ribosomal proteins and nucleotides in 18S rRNA cross-linked to 4-thioU or 6-thioG at [−3] or [+4] of mRNA in 48S complexes mapped onto corresponding ribosomal proteins and regions of 16S rRNA (gray) in the crystal structure of a complex of mRNA (blue) and the T. thermophilus 30S subunit (Yusupova et al. 2001). T. thermophilus rpS19 and rpS7 correspond to eukaryotic rpS15 and rpS5 and are red and magenta, respectively. Other ribosomal proteins are light blue. The mRNA nucleotides at [+4] and [−3] are red and magenta, respectively. Positions of nucleotides in 16S rRNA that correspond to C1696 and AA1818–1819 of 18S rRNA are shown as green and red spheres, respectively.
To identify the approximate region of cross-linking of [−3] and [+4] nucleotides to 18S rRNA, it was extracted after irradiation of 48S complexes, hybridized with DNA oligonucleotides complementary to different regions, digested with RNase H, and separated by electrophoresis. Attribution of individual 32P-labeled UV-cross-linked fragments of 18S rRNA took into account their reduced mobility due to covalently linked mRNA. The exact cross-linked nucleotide was identified by primer extension.
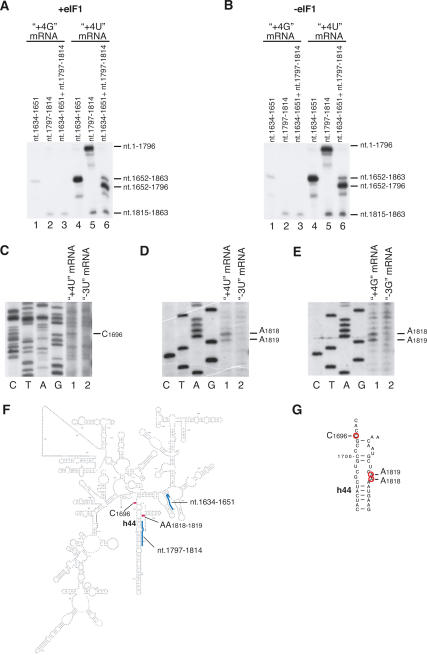
As with cross-linking to ribosomal proteins, cross-linking of 18S rRNA to the [+4] nucleotide was identical in 48S complexes assembled with or without eIF1, and thioG cross-linked less efficiently than thioU (Fig. 3A,B). In contrast to cross-linking of ribosomal proteins, cross-linking of thioU and thioG at [+4] to 18S rRNA differed significantly. Both nucleotides cross-linked to nucleotides 1652–1863, but further analysis showed that thioG[+4] cross-linked exclusively to nucleotides 1815–1863 (Fig. 3A,B, lanes 3), whereas thioU[+4] cross-linked to this region weakly but cross-linked strongly to nucleotides 1652–1796 (Fig. 3A,B, lanes 6). Cross-linking sites were then determined precisely: ThioU[+4] cross-linked mostly to C1696 and to some extent to AA1818–1819, whereas thioG[+4] cross-linked only to AA1818–1819 (Fig. 3C–E). In control experiments (Fig. 3C–E, lanes 2), primer extension was done on 48S complexes assembled on mRNAs containing thioU or thioG at [−3]. The positions of C1696 and AA1818–1819 in h44 of 18S rRNA are shown on the secondary structure of 18S rRNA (Fig. 3F,G) and are mapped onto the corresponding nucleotides of 16S rRNA in the crystal structure of the T. thermophilus 30S subunit (Fig. 4). In conclusion, in 48S complexes, U[+4] in mRNA specifically cross-linked to C1696 and to some extent to AA1818–1819, whereas G[+4] cross-linked exclusively to AA1818–1819. U and G both also cross-linked to rpS15.
Figure 3.
UV cross-linking of 4-thioU and 6-thioG at position +4 of mRNA with 18S rRNA in 48S complexes. (A,B) RNase H digestion of 18S rRNA cross-linked to 32P-labeled (CAA)n-AUG-(CAA)m mRNA derivatives containing 4-thioU or 6-thioG at [+4] in 48S complexes assembled with (A) or without (B) eIF1. 18S rRNA was digested in the presence of DNA primers complementary to nucleotides 1634–1651 and 1797– 1814, as indicated, and analyzed by electrophoresis in denaturing 12% PAGE and autoradiography. 18S rRNA fragments to which 4-thioU or 6-thioG at [+4] of mRNA had cross-linked in 48S complexes are shown on the right. (C–E, lanes 1) Determination of exact sites of cross-linking of (CAA)n-AUG-(CAA)m mRNA derivatives containing 4-thioU or 6-thioG at [+4] to 18S rRNA in 48S complexes by primer extension analysis. (Lanes 2) In control reactions UV cross-linking was done with 48S complexes assembled on (CAA)n-AUG-(CAA)m mRNA derivatives containing 4-thioU or 6-thioG at [−3]. The positions of RT stop sites are indicated on the right. Lanes C, T, A, and G depict 18S rRNA sequence generated using the same primer. (F) Secondary structure of rabbit 18S rRNA. Positions of cross-linked nucleotides and primers used for RNase H digestion are shown as red and blue bars, respectively. (G) Part of helix 44 of 18S rRNA showing cross-linked nucleotides (red circles).
Contacts of the nucleotide at position [−3] of mRNA with components of the 48S complex
Unlike the [+4] nucleotide, thioU or thioG at [−3] did not cross-link to 18S rRNA, but thioU at [−3] cross-linked specifically with equal efficiency to two proteins whereas thioG cross-linked only to the larger one (Fig. 2A,B, lanes 2,4). As for the [+4] nucleotide, cross-linking was identical in 48S complexes formed with or without eIF1 and was less efficient with thioG than thioU. The size of the smaller protein (∼21 kDa) indicated that it was a ribosomal protein. Taking into consideration the “northwest” shift of cross-linked proteins in 2D gels, we attributed cross-linking of U[−3] to rpS5 (Fig. 2E,F). Its identity was confirmed by mass-spectrometry sequencing of QAVDVFPLR and TIAEC*LADELINAAK peptides. It is a homolog of prokaryotic rpS7, shown on the crystal structure of the T. thermophilus 30S subunit (Fig. 4). The ∼38-kDa molecular weight of the larger protein indicated that it could be eIF2α or a subunit of eIF3. To identify it, we assembled 48S complexes using eIF2 with a truncated α-subunit (“ΔeIF2α”) from HeLa cells (Fig. 2G) and then exploited the observation that eIF5-induced hydrolysis of eIF2-bound GTP in 48S complexes releases eIF2 but not eIF3 (Unbehaun et al. 2004). The N-terminal sequence of ΔeIF2α (PGLS, identical to that of intact eIF2α) and its mobility in SDS-PAGE indicated that it was C-terminally truncated by 1.5–2 kDa; such cleavage is mediated by caspases (Satoh et al. 1999). eIF2 containing ΔeIF2α was ∼30% as active in 48S complex formation as intact eIF2 (data not shown). The lower activity may be due to the substoichiometric amount of ΔeIF2α in eIF2 compared with eIF2 with intact eIF2α (Fig. 2G). The ∼38-kDa protein cross-linked to thioU and thioG at [−3] was identified as eIF2α by cross-linking 48S complexes assembled with eIF2 containing ΔeIF2α on mRNA with thioU[−3] (which yielded a cross-linked protein with altered mobility) and by cross-linking 48S complexes after incubation with eIF5 (which led to specific loss of this band) (Fig. 2H, lanes 2,3). Cross-linking of eIF2α to [−3] was specific: No cross-linking was observed to [−4] and only very little to [−2] (V.G. Kolupaeva, A.V. Pisarev, C.U.T. Hellen, and T.V. Pestova, in prep.). In conclusion, in 48S complexes, thioU[−3] in mRNA cross-links specifically to rpS5 and eIF2α, whereas thioG[−3] cross-links exclusively to eIF2α.
48S complex formation on CAA-GUS mRNAs with upstream AUGs in different nucleotide contexts in the presence of α-subunit- and β-subunit-deficient mammalian eIF2
UV cross-linking data showed that G residues at [+4] and [−3] in mRNA specifically bound AA1818–1819 of 18S rRNA and eIF2α, respectively. To prove eIF2α’s functional role in recognizing initiation codon context, we compared 48S complex assembly using complete eIF2, eIF2 lacking either eIF2α (eIF2βγ) or eIF2β (eIF2αγ), or eIF2βγ with recombinant eIF2α on (CAA)n-AUGbad/bad-GUS, (CAA)n-AUGbad/good-GUS, and (CAA)n-AUGgood/bad-GUS mRNAs. These mRNAs have an unstructured 5′-UTR lacking potential near-cognate initiation codons and contain additional AUG triplets in different contexts upstream of the GUS initiation codon (Fig. 5A; Pestova and Kolupaeva 2002). The AUGbad/bad triplet contained [−3] and [+4] pyrimidines, AUGbad/good had a [−3] pyrimidine and a [+4] purine, and AUGgood/bad had a [−3] purine and a [+4] pyrimidine. Small quantities of eIF2βγ and eIF2αγ (Fig. 5B, lanes 2,3) were obtained using standard eIF2 purification procedures (Anthony et al. 1990; Materials and Methods). eIF1 is the principal factor that allows recognition of initiation codon context by scanning 43S complexes, so 48S complexes were assembled with and without eIF1. 48S complex formation did not depend on when eIF1 was added, and in all experiments described in this section identical data were obtained if eIF1 was added simultaneously with other translation components or if 48S complexes were first assembled without eIF1 and were then incubated with eIF1 for 15 min more. Consistent with our previous report (Pestova and Kolupaeva 2002), ∼90% of 43S complexes assembled with eIF1 and complete eIF2 scanned to the GUS initiation codon of (CAA)n-AUGbad/bad-GUS mRNA, whereas in the absence of eIF1, 48S complexes assembled mostly on the first AUGbad/bad triplet despite its poor context (Fig. 5C, lanes 2,3). 48S complexes formed with eIF2αγ just as with complete eIF2 (Fig. 5C, lanes 4,5). No differences in 48S complex formation were detected on (CAA)n-AUGbad/good-GUS and (CAA)n-AUGgood/bad-GUS mRNAs in the presence of eIF2αγ or complete eIF2 (data not shown). Consistently, there was no difference in cross-linking of eIF2α to thioU or thioG at [−3] in 48S complexes formed with complete eIF2 or eIF2αγ (Fig. 5D, lanes 1,2; data not shown).
Figure 5.
Activities of α-subunit- and β-subunit-deficient eIF2 in 48S complex formation. (A) Sequence of the 5′-UTR of (CAA)n-AUGbad/bad-GUS, (CAA)n-AUGgood/bad-GUS, and (CAA)n-AUGbad/good-GUS mRNAs with initiation codons in bold. Context residues −3 to +4 are underlined. (B) Coomassie-stained SDS-PAGE with resolved complete, α-subunit- and β-subunit-deficient eIF2, and recombinant eIF2α. The positions of eIF2 subunits are indicated on the right. (C,F,G) Toe-print analysis of 48S complexes assembled on mRNAs (shown in A) from 40S subunits, Met-tRNAMeti, complete, α-subunit- and β-subunit-deficient eIF2, and other eIFs as indicated. Full-length cDNAs and toe-prints due to 48S complexes are shown on the side. (D,E) UV cross-linking of 32P-labeled (CAA)n-AUG-(CAA)m mRNA derivatives containing 4-thioU or 6-thioG at [−3] in 48S complexes assembled using forms of eIF2 as indicated, assayed by SDS-PAGE and autoradiography. eIF2α and rpS5 are indicated on the right.
The role of eIF2α in 48S complex formation was investigated using (CAA)n-AUGbad/good-GUS and (CAA)n-AUGgood/bad-GUS mRNAs. This combination of mRNAs containing upstream AUG triplets with only one purine at either [−3] or [+4] was optimal for these studies because 48S complex formation on the first AUG triplet of (CAA)n-AUGbad/bad-GUS mRNA in the presence of eIF1 is inefficient even with complete eIF2, and quantitating possible reductions in 48S complex formation with eIF2βγ reliably would be difficult. On the other hand, 48S complex formation on the first AUG of (CAA)n-AUGgood/good-GUS mRNA would be too efficient to permit detection of potential leaky scanning. In addition, (CAA)n-AUGbad/good-GUS and (CAA)n-AUGgood/bad-GUS mRNAs allow the relative effects of purines at different positions on the efficiency of 48S complex formation to be compared. With complete eIF2 and without eIF1, 48S complexes formed almost exclusively on the first AUG triplet on both mRNAs (Fig. 5F,G, lanes 5). In the presence of eIF1, complex formation on the first AUG triplet with a [−3] purine was more efficient, constituting ∼80% of 48S complexes whereas 48S complex formation on the first AUG with a [+4] purine constituted ∼50% of the total (Fig. 5F,G, lanes 6). The [−3] purine was therefore relatively more important for these mRNAs than that at [+4]. In the absence of eIF1, total 48S complex formation on the two AUGs for both mRNAs was only 10% lower with eIF2βγ than with complete eIF2. Initiation on both mRNAs was slightly leakier: 10%–15% of 48S complexes formed on the GUS AUG with eIF2βγ, whereas only ∼4% of 48S complexes formed there with complete eIF2 (Fig. 5F,G, lanes 1,5). Although it had only a minor effect in the absence of eIF1, the lack of eIF2α strongly affected 48S complex formation in eIF1’s presence. Total 48S complex formation with eIF2βγ and eIF1 on the two AUG triplets of both mRNAs was reduced threefold (Fig. 5F,G, cf. lanes 2,6). The relative reduction in 48S complex formation on the first AUG triplet was higher with (CAA)n-AUGgood/bad-GUS mRNA, in which case the ratio between 48S complex formation on the first and second AUG triplets fell to 1:1 from 4:1 in the presence of complete eIF2 (Fig. 5G, lanes 2,6) and became similar to the ratio of 48S complex formation on the first (with an unfavorable [−3] pyrimidine) and second AUGs of (CAA)n-AUGbad/good-GUS mRNA (Fig. 5F, lanes 2,6). This result suggests that in the absence of eIF2α, 43S complexes cannot sense the nature of the [−3] nucleotide. The similar relative efficiencies of 48S complex formation on the two AUG triplets on both mRNAs despite the first AUG triplet of (CAA)n-AUGbad/good-GUS mRNA having a favorable [+4] purine suggest that the “+4 nucleotide rule” might be secondary to the “−3 nucleotide rule” and may not function efficiently in the absence of the eIF2α/[−3] nucleotide interaction. The fact that 48S complex formation with eIF2α-deficient eIF2 was strongly reduced on both AUGbad/good and AUGgood/bad suggests that interaction of eIF2α with the [−3] nucleotide is, irrespective of its nature, generally important for resistance of 48S complexes to dissociation by eIF1. Addition of recombinant eIF2α to reaction mixtures containing eIF2βγ restored the efficiency of 48S complex formation on both AUG codons to the level observed with complete eIF2 (Fig. 5F,G, lanes 2,4,6). Consistently, recombinant eIF2α added to reaction mixtures with eIF2βγ was cross-linked to thioU or thioG at [−3] in 48S complexes (Fig. 5D [lanes 3,4], E). 43S complexes became slightly less leaky and fewer 48S complexes assembled on the GUS AUG codon with eIF2βγ and recombinant eIF2α than with native complete eIF2 (Fig. 5F,G, lanes 4,6). The eIF2α N-terminal domain may interact with the [−3] nucleotide, in which case the N-terminal tag may influence this interaction, but we were reluctant to tag the C terminus of eIF2α because its C-terminal domain interacts with eIF2γ. In conclusion, these data suggest that interaction of eIF2α with the [−3] nucleotide is generally important for 48S complex formation in the presence of eIF1, and that eIF2α likely interacts more strongly with a purine at [−3], protecting 48S complexes more from dissociation by eIF1.
Interaction of eIF2α with the [−3] nucleotide of mRNA in 48S complexes after eIF5-induced hydrolysis of eIF2-bound GTP
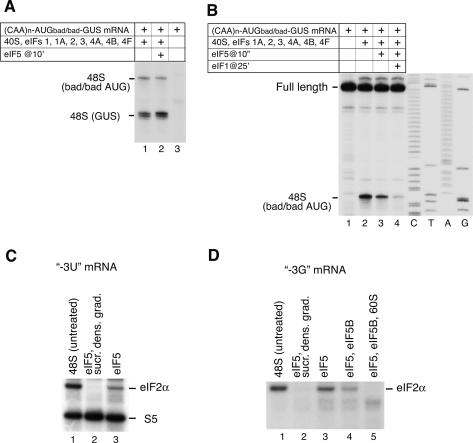
Our data suggest that eIF2α’s interaction with the [−3] nucleotide stabilizes 48S complexes against dissociation by eIF1. However, it is generally accepted that eIF5-induced hydrolysis of eIF2-bound GTP leads eIF2 to dissociate from 48S complexes. If eIF1 can dissociate aberrant initiation complexes after GTP hydrolysis, then one would expect that if ribosomal subunit joining does not occur immediately after this event, then even 48S complexes assembled on AUG triplets with a [−3] purine would be dissociated at this stage. However, 48S complexes formed with eIF1 on the good context AUG codon of the GUS ORF of (CAA)n-AUGbad/bad-GUS mRNA remained intact after 15 min incubation with eIF5 (Fig. 6A, lanes 1,2). This result was not due to a hypothetical inability of eIF1 to discriminate the context of the initiation codon after hydrolysis of eIF2-bound GTP, because ∼95% of 48S complexes formed on the first bad context AUG of the same mRNA without eIF1 could still be dissociated by eIF1 after incubation with eIF5 (Fig. 6B, lanes 2,4). The upstream AUG triplet had poor context at [−3] and [+4] whereas the downstream AUG triplet had good context at both positions. The [+4] purine could conceivably be sufficient to stabilize 48S complexes after hydrolysis of eIF2-bound GTP. However, retaining a purine only at [−3] of the first AUG codon in (CAA)n-AUGgood/bad-GUS mRNA yielded 48S complexes that were as resistant to eIF1-mediated dissociation after GTP hydrolysis as eIF5-untreated 48S complexes (data not shown). To account for this result, we tested if interaction of the [−3] purine switches from eIF2α to another component of the 48S complex after GTP hydrolysis, which could render 48S complexes resistant to eIF1-mediated dissociation in the absence of eIF2. Just as for eIF5-untreated 48S complexes, no specific interaction was detected between thioG[−3] and 18S rRNA or any ribosomal protein after eIF5-induced hydrolysis of eIF2-bound GTP (Fig. 6D; data not shown).
Figure 6.
Influence of eIF5-induced hydrolysis of eIF2-bound GTP on 48S complex formation on AUG triplets in good and bad context (A,B) and on UV cross-linking of eIF2α to the −3 nucleotide of mRNA in 48S complexes (C,D). (A,B) Toe-print analysis of 48S complexes assembled on (CAA)n-AUGbad/bad-GUS mRNA from 40S subunits, Met-tRNAMeti, and eIFs as indicated. Toe-prints due to 48S complexes are shown on the left. (C,D) UV cross-linking of 32P-labeled (CAA)n-AUG-(CAA)m mRNA derivatives containing 4-thioU or 6-thioG at [−3] with components of 48S complexes before and after incubation with eIF5, eIF5B, and 60S subunits, as indicated. In lanes 2 48S complexes incubated with eIF5 were subjected to sucrose density gradient centrifugation before UV cross-linking. Cross-linked proteins were assayed by SDS-PAGE and autoradiography. eIF2α and rpS5 are indicated on the right.
Whereas the affinity to aminoacylated tRNA of elongation factor EF-Tu/GTP and EF-Tu/GDP-bound differs by a factor of 104, the affinity of yeast eIF2-GDP to Met-tRNAMeti is only ∼20-fold lower than of eIF2-GTP: This small difference might lead to incomplete dissociation of eIF2 from 40S subunits upon GTP hydrolysis (Kapp and Lorsch 2004). Although eIF2 dissociated entirely from 40S subunits in our experiments (Fig. 2H, lane 3; Unbehaun et al. 2004), they all included sucrose density gradient centrifugation of 48S complexes after eIF5-induced hydrolysis of GTP prior to analysis of eIF2’s association with 40S subunits. To determine whether the stringency of sucrose density gradient centrifugation dissociated eIF2 from 40S subunits in these experiments, we assayed its influence on cross-linking of thioU and thioG at [−3] in mRNA in eIF5-treated 48S complexes. As expected, no cross-linking to eIF2α was observed with either mRNA if eIF5-treated 48S complexes were subjected to sucrose density gradient centrifugation before irradiation (Fig. 6C,D, lanes 1,2). However, if this step was omitted, after GTP hydrolysis 30%–35% and 85%–90% of eIF2 still cross-linked to thioU and to thioG at [−3], respectively (Fig. 6C,D, lane 3). This indicates that hydrolysis of eIF2-bound GTP does not cause complete dissociation of eIF2 from 48S complexes. Moreover, the extent of eIF2 release depends on the nature of the [−3] nucleotide and is much lower when it is a purine. Addition of eIF5B to a reaction mixture with eIF5 almost completely abrogated cross-linking of eIF2α to thioU[−3] and reduced cross-linking to thioG[−3] by 70% (Fig. 6D, lane 4; data not shown). These results suggest that eIF5B promotes dissociation of eIF2 from the 40S subunit after hydrolysis of eIF2-bound GTP, which is nevertheless not complete if the mRNA has a [−3] purine. The absence of UV cross-linking of eIF2α to either thioU or thioG at [−3] after treatment of 48S complexes with eIF5, eIF5B, and 60S subunits (Fig. 6D, lane 5; data not shown) indicated complete conversion of 48S complexes into 80S ribosomes and confirmed that incubation with eIF5 alone or together with eIF5B in identical conditions (Fig. 6D, lanes 3,4) led to complete hydrolysis of eIF2-bound GTP. In case some eIF1 was lost from 48S complexes during their initial purification by sucrose density gradients, we compared the effect of adding eIF5 alone or together with eIF1–48S complexes on UV cross-linking of eIF2α to thioG[−3]: No difference was detected, which means that eIF2 release was not affected (data not shown).
eIF5-induced hydrolysis of eIF2-bound GTP, therefore, does not completely dissociate eIF2 from 48S complexes, and the fact that the nature of the [−3] nucleotide influences eIF2 release suggests that mRNA stabilizes binding of eIF2–48S complexes after GTP hydrolysis through interaction of eIF2α with the [−3] nucleotide. The fact that only a small fraction of eIF2 was released from 48S complexes assembled on AUG codons with a [−3] purine upon hydrolysis of eIF2-bound GTP could account for resistance of these complexes to dissociation by eIF1 even after treatment with eIF5. Interaction of eIF2α with the [−3] purine likely also contributes to the stability of 48S complexes after GTP hydrolysis even in the absence of eIF1, because in this case treatment with eIF5 of 48S complexes assembled on a bad context AUG codon led to 65% dissociation (Fig. 6B, lane 3). Association of eIF2 with 48S complexes after hydrolysis of eIF2-bound GTP might also prevent Met-tRNAMeti from dissociating from 40S subunits before ribosomal subunit joining.
Discussion
eIF1’s position on the 40S subunit between the platform and initiator tRNA suggests that it acts indirectly to ensure the fidelity of initiation codon selection and, specifically, to enable 43S complexes to discriminate against AUG triplets in suboptimal context (Pestova and Kolupaeva 2002; Lomakin et al. 2003). The finding that the C-terminal domain of prokaryotic IF3 (which is not homologous to eIF1) can bind the same region of the 40S subunit and perform many of eIF1’s functions in initiation codon selection, including enabling 43S complexes to recognize initiation codon context, also favors an indirect mode of action for eIF1 (Lomakin et al. 2006). Our hypothesis that eIF1 acts by antagonizing conformational changes in the 48S complex that occur as a result of initiation codon recognition and base-pairing with the anticodon suggests that the role of the key −3 and +4 context nucleotides is to stabilize such changes by interacting with components of the 48S complex. Here, we used UV cross-linking to characterize and compare the specificity of interactions of thioU and thioG at these positions with constituents of this complex. In a separate study, we used mRNAs containing single thioU residues at positions −26 to +11 to map the mRNA path on the 40S subunit in 48S complexes (V.G. Kolupaeva, A.V. Pisarev, C.U.T. Hellen, and T.V. Pestova, in prep.). Its similarity to the mRNA path on the 70S ribosome as determined by crystallography (Yusupova et al. 2001) justifies using the structure of the mRNA/30S subunit complex to model our cross-linking data.
UV cross-linking in 48S complexes formed with and without eIF1
IF3, a functional analog of eIF1, alters the position of mRNA on 30S subunits (La Teana et al. 1995; Shapkina et al. 2000), so we assayed interactions of the −3 and +4 nucleotides in 48S complexes assembled with and without eIF1. The interactions of thioU or thioG at both positions were unaffected by eIF1. Even if eIF1 influences the positions of mRNA or Met-tRNAMeti in scanning ribosomal complexes, the final conformation of 48S complexes with established codon–anticodon base-pairing appears not to depend on eIF1’s involvement in their assembly. We detected eIF1 in 48S complexes after eIF5-induced hydrolysis of eIF2-bound GTP (Unbehaun et al. 2004), but the observation that eIF1 was released from minimal yeast initiation complexes following codon–anticodon base-pairing (Maag et al. 2005) suggests that in mammalian 48S complexes, eIF1 might be displaced from its original location on the 40S subunit but be retained in these complexes by interaction with eIF3. If this is so, the apparently identical position of mRNA in 48S complexes assembled with and without eIF1 is not surprising.
UV cross-linking to the [+4] position
Both thioU[+4] and thioG[+4] cross-linked to rpS15. However, whereas thioU cross-linked weakly to AA1818–1819 and strongly to C1696 in h44 of 18S rRNA, thioG cross-linked exclusively to AA1818–1819. Specific mRNA cross-linking to components of the 48S complex has not previously been analyzed, so we compared our data with mRNA cross-linking in eukaryotic 80S complexes phased by cognate tRNA and in prokaryotic 70S complexes. Cross-linking of thioU[+4] to rp15 in 48S complexes was consistent with the same interaction in 80S complexes (Bulygin et al. 2005). rpS19, the prokaryotic homolog of rpS15, is located in the head of the 30S subunit (Fig. 4; Wimberly et al. 2000). Its C-terminal tail points toward the interface side but does not reach the A-site codon, so cross-linking of rpS15 is likely due to N- or C-terminal extensions relative to prokaryotic rpS19.
Cross-linking of mRNA to AA1818–1819 has been detected with midrange nucleotide derivatives but not with “zero-length” cross-linkers: No cross-linking of AA1818–1819 to thioU[+4] was observed in phased or unphased 80S complexes (Demeshkina et al. 2000; Bulygin et al. 2005). The equivalent prokaryotic nucleotides (AA1492–1493 in T. Thermophilus) flip out upon binding of cognate aminoacyl tRNA to the A-site during elongation and interact with the minor groove of the first two base pairs of the base-paired codon–anticodon helix, thereby monitoring the fidelity of elongator tRNA selection (Ogle et al. 2001). Flipping out of AA1492–1493 also occurs during prokaryotic initiation when IF1 binds to the A-site area of the 30S subunit; these bases splay apart whereas they stack together when cognate tRNA binds to the A-site (Carter et al. 2001). Binding of eIF1A, the eukaryotic IF1 homolog (Battiste et al. 2000) or other factors to the 40S subunit might also alter the conformation of the upper part of h44 and flip out AA1818–1819. Such conformational changes could account for “zero-length” cross-linking of thioU and thioG at [+4] to AA1818–1819 in 48S but not 80S complexes. Cross-linking of thioU[+4] to C1696 in 48S complexes was not consistent with cross-linking of thioU[+4] to the equivalent of rabbit C1691 in H28 of 18S rRNA in human 80S complexes (Bulygin et al. 2005). This discrepancy cannot be explained by the difference in positions of mRNA in 48S and 80S complexes: In our recent experiments C1691 cross-linked specifically to thioU[+8] in 48S and 80S complexes (V.G. Kolupaeva, A.V. Pisarev, C.U.T. Hellen, and T.V. Pestova, in prep.), consistent with cross-linking of thioU[+8] to the equivalent nucleotide (C1395) in prokaryotic 70S complexes (Rinke-Appel et al. 1993). In prokaryotes, C1400 and AA1492–1493 (equivalents of rabbit C1696 and AA1818–1819) are opposite each other, flanking the mRNA (Fig. 4). Cross-linking of thioU to both sites suggests that structural rearrangements in 48S complexes cause them to be closer to each other than their equivalents in prokaryotic 30S subunit/70S ribosome crystal structures. The inability of thioG to cross-link to C1696 might be due to its specific interaction with A1818 and/or A1819, which could cause further structural adjustments that preclude this cross-link.
UV cross-linking to the [−3] position
In 48S complexes, neither thioU nor thioG at [−3] cross-linked efficiently to 18S rRNA, but thioU[−3] cross-linked to rpS5 and eIF2α and thioG[−3] cross-linked only to eIF2α. Cross-linking of rpS5 to the [−3] position is consistent with the similar paths of mRNA on eukaryotic 40S and prokaryotic 30S subunits: In prokaryotic ribosomal complexes, thioU[−3] cross-links specifically to rpS7, a homolog of eukaryotic rpS5 (Fig. 4; La Teana et al. 1995). Cross-linking of the [−3] nucleotide to rpS5 was also observed in eukaryotic 80S complexes (Demeshkina et al. 2003); this study also reported its cross-linking to rpS26 (and to rpS2 and rpS3, which we suggest was nonspecific, taking into account the positions of prokaryotic analogs of eukaryotic rpS2 and rpS3 in the 30S subunit; Wimberly et al. 2000). In contrast to the study by Demeshkina et al. (2003), we saw specific cross-linking of rpS26 to mRNA in 48S complexes only to thioU at [−8] to [−11] but not at all at [−3] (V.G. Kolupaeva, A.V. Pisarev, C.U.T. Hellen, and T.V. Pestova, in prep.).
The mRNA path in 48S complexes has not been studied, so specific cross-linking of eIF2α to [−3] in mRNA has not been reported. eIF2α consists of structured N-terminal and C-terminal domains that are mobile relative to each other; the latter binds eIF2γ (Yatime et al. 2004). eIF2α might thus bind the [−3] nucleotide either through the N-terminal domain or through its unstructured C-terminal tail. The absence of the ∼10 C-terminal amino acids of eIF2α and, interestingly, of eIF2β did not influence this interaction. Consistent with the affinities to Met-tRNAMeti of eIF2-GTP and eIF2-GDP differing by only one order of magnitude (Kapp and Lorsch 2004), in the absence of sucrose density gradient centrifugation, eIF5-induced hydrolysis of eIF2-bound GTP did not lead to complete dissociation of eIF2 from 48S complexes so that 30%–35% and 85%–90% of eIF2α could still cross-link to thioU[−3] and thioG[−3], respectively. The fact that the nature of the [−3] nucleotide influenced its cross-linking to eIF2α after eIF5-induced GTP hydrolysis suggests that the eIF2–mRNA interaction influences release of eIF2 during subunit joining. It is possible that without this interaction, GTP hydrolysis would result in greater and even complete eIF2 dissociation.
The finding that eIF5B enhances release of eIF2 from 48S complexes after GTP hydrolysis merits special attention. Although unlike its prokaryotic homolog IF2, binding of eIF5B to Met-tRNAMeti has not been shown directly, this interaction might occur on the 40S subunit and after binding to 48S complexes, eIF5B might compete with eIF2 for interaction with Met-tRNAMeti. Weakening of eIF2/Met-tRNAMeti binding after hydrolysis of bound GTP could permit an interaction between Met-tRNAMeti and the C-terminal domain IV of eIF5B to be established, and consequently promote release of eIF2. However, complete release of eIF2 from 48S complexes assembled on mRNA containing thioG[−3] occurred only after ribosomal subunit joining, which suggests that eIF2 is completely released only during the actual ribosomal subunit joining event promoted by eIF5B. mRNA, therefore, influences release of eIF2 as well as of eIF3 from initiation complexes (Unbehaun et al. 2004).
Activities of α-subunit- and β-subunit-deficient eIF2 in 48S complex formation
Specific UV cross-linking to thioG[−3] in mRNA in 48S complexes suggests that eIF2α is involved in recognition of initiation codon context and thus in initiation codon selection. The functionality of this interaction was confirmed in experiments on 48S complex formation in the presence of eIF2α-deficient eIF2βγ on two mRNAs, both containing two AUG triplets, of which the first had a purine residue either at [−3] or at [+4]. With complete eIF2 but without eIF1, 48S complexes formed almost exclusively on the first AUG triplets of both mRNAs, but in the presence of eIF1, 48S complex formation was more efficient on the AUG triplet with the [−3] purine (80% of total 48S complexes) than with the [+4] purine (50% of total 48S complexes). In the absence of eIF1, the lack of eIF2α had little effect on the efficiency or specificity of 48S complex formation so that 43S complexes stopped efficiently on the first AUG triplet irrespective of its context. In eIF1’s presence, the lack of eIF2α strongly influenced 48S complex formation. First, the combined efficiency of 48S complex formation on two AUG triplets on both mRNAs was threefold lower than with complete eIF2. Second, whereas the ratio of 48S complexes formed on the first AUG triplet with a [−3] purine and on the second AUG triplet was 4:1 in the presence of complete eIF2, it fell to 1:1 in the absence of eIF2α and became similar to the ratio of 48S complex formation on mRNA with two AUG triplets in which the first was flanked by a [−3] pyrimidine. In the presence of eIF1, 43S complexes assembled without eIF2α therefore could not sense the nature of the [−3] nucleotide and 48S complexes formed with equal efficiency on AUG triplets whether there was a purine or a pyrimidine at [−3]. This result confirmed the suggested role for eIF2α in discriminating the [−3] context nucleotide. The reduced efficiency of 48S complex formation on the AUG triplet with a [−3] pyrimidine in the absence of eIF2α also suggests that eIF2α’s interaction with the [−3] nucleotide, irrespective of its nature, is generally important for 48S complex formation in the presence of eIF1 but that it is the strength of interaction (which is higher for purines) that is responsible for the [−3] context rule. Our finding that eIF2 is not fully released from 48S complexes upon eIF5-induced GTP hydrolysis and that the extent of its release depends on the nature of the [−3] nucleotide (being only 10%–15% with G at this position) could account for the resistance of 48S complexes to eIF1-mediated dissociation after hydrolysis of eIF2-bound GTP and before the ribosomal subunit joining step.
Although interaction of the [+4] nucleotide with rpS15 was not base-specific and rpS15 is also cross-linked to thioU[+5] (V.G. Kolupaeva, A.V. Pisarev, C.U.T. Hellen, and T.V. Pestova, in prep.), we cannot exclude the possibility that the rpS15-[+4] nucleotide interaction is important for initiation codon selection. We cannot directly test the functional importance of interaction of the [+4] nucleotide with AA1818–1819, but the base specificity of this interaction points to the fact that AA1818–1819 are involved not only in monitoring the fidelity of elongator tRNA selection, but also in selection of the initiation codon during initiation. By analogy with eIF2α, the interaction of the [+4] nucleotide with components of the 48S complex (AA1818–1819 and/or rp S15) might also be generally important to stabilize 48S complexes assembled on AUG triplets whether they have a purine or a pyrimidine at [+4].
Materials and methods
Plasmids
Vectors for expression of His6-tagged eIF1, eIF1A, eIF4A, eIF4B, eIF5, and Escherichia coli methionyl-tRNA synthetase, and for (CAA)n-AUGbad/bad-GUS mRNA transcription have been described (Pestova et al. 1996, 1998, 2000; Pestova and Kolupaeva 2002; Lomakin et al. 2006). The bovine eIF2α coding region was amplified by PCR from pUKC50 (Green et al. 1991) and cloned between BamH1 and HindIII restriction sites of pET28b (Novagen) yielding pET(His6-eIF2α). Transcription vectors for (CAA)n-AUG-(CAA)m mRNA derivatives containing U or G at [−3] or [+4] were made by inserting complementary oligonucleotides corresponding to a T7 promoter, 5′-UTR, coding region, and SmaI restriction site between the BamH1 and HindIII sites of pBR322. (CAA)n-AUGbad/good-GUS and (CAA)n-AUGgood/bad-GUS transcription vectors were made using the same strategy as for the (CAA)n-AUGbad/bad-GUS vector (Pestova and Kolupaeva 2002).
In vitro transcription
All mRNAs were transcribed using T7 RNA polymerase. (CAA)n-AUGbad/bad-GUS, (CAA)n-AUGgood/bad-GUS, and (CAA)n-AUGbad/good-GUS mRNAs were transcribed as described (Pestova and Kolupaeva 2002). For UV cross-linking experiments, 32P-labeled derivatives of (CAA)n-AUG-(CAA)m mRNA containing 4-thioU or 6-thioG (8 × 106 c.p.m./μg) were transcribed from SmaI-digested plasmids in the presence of 4-thioUTP (Ambion) or 6-thioGTP (Jena Bioscience), [α32P]-CTP (222 Tbq/mmol), and a rAC transcription primer (Dharmacon). For toe-printing experiments, derivatives of (CAA)n-AUG-(CAA)m mRNA were transcribed from EagI-digested plasmids.
Purification of factors and ribosomal subunits, and aminoacylation of initiator tRNA
40S and 60S subunits, eIF2, eIF3, eIF4F, and eIF5B were purified from rabbit reticulocyte lysate (RRL) and recombinant eIF1, eIF1A, eIF4A, eIF4B, eIF5, and E. coli methionyl-tRNA synthetase were expressed in E. coli BL21(DE3) and purified as described (Pestova et al. 1996, 1998, 2000; Lomakin et al. 2006). α-Subunit-deficient eIF2 was purified as described (Anthony et al. 1990). β-Subunit-deficient eIF2 is always obtained in small amounts during eIF2 purification from RRL (Pestova et al. 2000) as a peak eluted from MonoQ two fractions earlier than complete eIF2. eIF2 with a truncated α-subunit was purified in small quantities from HeLa cells using the purification procedure previously described for eIF2 from RRL (Pestova et al. 2000) as a peak eluted from MonoQ slightly earlier than complete eIF2. Recombinant eIF2α was expressed in E. coli BL21(DE3) and purified on Ni2+-NTA (Qiagen) and MonoQ. Total native rabbit tRNA (Novagen) was aminoacylated by recombinant methionyl-tRNA synthetase as described (Pestova et al. 1996).
UV cross-linking experiments
48S complexes were assembled on 32P-labeled derivatives of (CAA)n-AUG-(CAA)m mRNA containing 4-thioU or 6-thioG at [−3] or [+4] by incubating 100 ng of mRNA, 10 pmol Met-tRNAMeti, 8 pmol 40S subunits, 5 μg of different forms of eIF2, 15 μg of eIF3, 2.5 of μg eIF4A, 0.5 μg of eIF4B, 2.5 μg of eIF4F, 0.2 μg of eIF1A, 0.2 μg of eIF1, and 3 μg of recombinant eIF2α (as indicated) in 100 μL of buffer A (20 mM Tris at pH 7.5, 100 mM KAc, 2 mM DTT, 2.5 mM MgAc2, 0.25 mM spermidine) containing 1 mM ATP, 0.4 mM GTPγS (or 0.4 mM GTP when eIF5 was included) for 10 min at 37°C. In reaction mixtures shown in Figures 2H (lane 3) and 6C,D (lanes 2) hydrolysis of eIF2-bound GTP was induced by incubating with 1 μg of eIF5 for 15 min. 48S complexes were purified by centrifugation in a Beckman SW55 rotor for 1h and 40 min at 4°C and 50,000 rpm in 10%–30% sucrose density gradients prepared in buffer A. [32P]-labeled mRNA in ribosomal fractions was monitored by Cherenkov counting. Equal amounts of counts (∼200,000 c.p.m.) of peak fractions were irradiated at 360 nm for 30 min on ice using a UV-Stratalinker (Stratagene) and used to identify cross-linked proteins and nucleotides in 18S rRNA. For experiments shown in Figure 6C (lane 3) and D (lanes 3,5), 48S complexes were purified by sucrose density gradient centrifugation, diluted threefold with buffer A + 0.4 mM GTP, incubated for 15 min at 37°C with 1 μg of eIF5, 1 μg of eIF5B, or 8 pmol 60S subunits, as indicated, and then subjected to UV irradiation.
Identification of cross-linked proteins
To identify UV-cross-linked eIFs, ∼20 μL of cross-linked ribosomal fractions containing equal amounts of counts were treated with RNase A and subjected to electrophoresis in NuPAGE 4%–12% Bis-Tris-Gel (Invitrogen) followed by autoradiography. UV-cross-linked ribosomal proteins were identified by acidic-SDS 2D gel electrophoresis. Complete cross-linked peak fractions (∼200,000 c.p.m.) were combined, transferred to buffer B (20 mM Tris-HCl at pH 7.5, 50 mM KCl, 2 mM MgCl2, 2 mM DTT, 0.1 mM EDTA), concentrated on microcon-YM10 centrifugal filter units (Millipore) to 100 μL of final volume, and treated with RNase A for 30 min at 37°C. These samples were combined with 100 μL of 40S subunits (OD260 = 100 o.u./mL) in buffer B. Proteins were extracted from these mixtures with 100 mM MgCl2 in 67% acetic acid and precipitated with acetone (Hardy et al. 1969). Samples were then resuspended in 8 M urea, 1% 2-mercaptoethanol, 10 mM bis-tris acetate (pH 4.2); incubated for 15 min at 37°C; and subjected to first-dimension electrophoresis (Yusupov and Spirin 1988) in 120-mm-long glass tubes with a 2.4-mm inner diameter. First-dimension gels were incubated for 10 min in cathode buffer and combined with second-dimension gels, which had been prepared as described (Schagger and von Jägow 1987). The separating gel (16.5% T and 3% C) contained 13.3% w/v glycerol. Gels were run for 12 h at 40 mA, stained with Simply Blue Safe Stain (Invitrogen), and destained with water for LC-nanospray tandem mass spectrometry of peptides derived by in-gel tryptic digestion at an in-house facility, or fixed with 10% methanol/5% glycerol for drying and autoradiography.
Identification of cross-linked nucleotides in 18S rRNA
After irradiating 48S complexes, rRNA, mRNA, and tRNA were phenol-chloroform extracted and ethanol precipitated. Regions of 18S rRNA cross-linked to 32P-labeled mRNA were first identified by RNase H digestion of 18S rRNA hybridized with a panel of ∼20-mer DNA oligonucleotides complementary to different regions of 18S rRNA essentially as described (Dontsova et al. 1992). 18S rRNA fragments were separated by electrophoresis in 12% denaturing PAGE. Cross-linked and uncross-linked 18S rRNA fragments were visualized by autoradiography and methylene blue staining, respectively. Cross-linked regions were identified and attributed to corresponding uncross-linked fragments of 18S rRNA on stained gels taking into account the reduced mobility of cross-linked rRNA fragments due to covalently bound 64-nt mRNA. Precise identification of cross-linked nucleotides in 18S rRNA was done by primer extension inhibition using primers 5′-CAAGTTCGACCGTCTTC-3′ and 5′-CC TTCCGCAGGTTCACC-3′ complementary to nucleotides 1783–1799 and 1840–1856 of 18S rRNA respectively, chosen on the basis of RNase H digestion.
Toe-printing analysis of 48S initiation complexes
Ribosomal 48S complexes assembled on (CAA)n-AUGbad/bad-GUS, (CAA)n-AUGgood/bad-GUS, and (CAA)n-AUGbad/good-GUS mRNAs, and derivatives of (CAA)n-AUG-(CAA)m mRNAs were analyzed by primer extension using AMV RT essentially as described (Pestova and Kolupaeva 2002). Reaction mixtures (40 μL) containing 2 pmol mRNAs, 5 pmol Met-tRNAMeti, 5 pmol 40S subunits, 1.5 μg of complete or subunit-deficient eIF2, 1.2 μg of eIF2α, 6 μg of eIF3, 1 μg of eIF4A, 0.2 μg of eIF4B, 1μg of eIF4F, 0.2 μg of eIF1, and 0.2 μg of eIF1A as indicated in the figure legends were incubated in buffer A (+1mM ATP + 0.4 mM GTP) at 37°C for 10 min, and in some cases for 15 min more with 1 μg of eIF5 or 1 μg of eIF5 and 0.3 μg of eIF1 (Fig. 6A,B). Toe-printing analysis was done using [32P]dATP and primers 5′-CATGACATTAACC-3′ and 5′-CGCGCTTTCCCACCAA CG-3′s complementary to pBR322 nucleotides 4307–4319 and GUS nucleotides 97–114, respectively. cDNA products were analyzed by electrophoresis through 6% polyacrylamide sequencing gel. PhosphorImager analysis was used to quantify the efficiency of initiation complex formation. All values presented in Results are the average of at least three independent experiments.
Acknowledgments
We thank R. Schneider and M. Tuite for generously providing antibodies against eIF2α and the bovine eIF2α clone, M. Yusupov for valuable technical advice, and A. Marintchev for helpful discussion. This work was supported by NIH grant R01 GM59660 (to T.V.P.) and R01 GM26796 (to W.C.M.).
Footnotes
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1397906
References
- Algire M.A, Maag D, Lorsch J.R. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Anthony D.D, Jr., Kinzy T.G, Merrick W.C. Affinity labeling of eukaryotic initiation factor 2 and elongation factor 1 α β γ with GTP analogs. Arch. Biochem. Biophys. 1990;281:157–162. doi: 10.1016/0003-9861(90)90426-y. [DOI] [PubMed] [Google Scholar]
- Battiste J.L, Pestova T.V, Hellen C.U, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- Bulygin K, Chavatte L, Frolova L, Karpova G, Favre A. The first position of a codon placed in the A site of the human 80S ribosome contacts nucleotide C1696 of the 18S rRNA as well as proteins S2, S3, S3a, S30, and S15. Biochemistry. 2005;44:2153–2162. doi: 10.1021/bi0487802. [DOI] [PubMed] [Google Scholar]
- Carter A.P, Clemons W.M, Brodersen D.E, Morgan-Warre R.J, Hartsch T, Wimberley B.T, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Cavener D.R, Ray S.C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3189. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A.M, Feng L, Donahue T.F. Met-tRNAMeti functions in directing the scanning ribosome to the start site of translation. Science. 1988;242:93–97. doi: 10.1126/science.3051379. [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Repkova M, Ven’yaminova A, Graifer D, Karpova G. Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P, and E sites: A crosslinking study with mRNA analogs carrying an aryl azide group at either the uracil or the guanine residue. RNA. 2000;6:1727–1736. doi: 10.1017/s1355838200000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeshkina N.A, Laletina E.S, Meshchaninova M.I, Repkova M.N, Ven’iaminova A.G, Graifer D.M, Karpova G.G. The mRNA codon environment at the P and E sites of human ribosomes deduced from photo crosslinking with pUUUGUU. Mol. Biol. 2003;37:147–155. [PubMed] [Google Scholar]
- Dontsova O, Dokudovskaya S, Kopylov A, Rinke-Appel J, Jünke N, Brimacombe R. Three widely separated positions in the 16S RNA lie in or close to the ribosomal decoding region; a site-directed cross-linking study with mRNA analogues. EMBO J. 1992;11:3105–3116. doi: 10.1002/j.1460-2075.1992.tb05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre A, Saintomé C, Fourrey J.L, Clivio P, Laugâa P. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid–protein interactions. J. Photochem. Photobiol. B. 1998;42:109–124. doi: 10.1016/s1011-1344(97)00116-4. [DOI] [PubMed] [Google Scholar]
- Green S.R, Fullekrug J, Sauer K, Tuite M.F. Isolation and characterisation of a bovine cDNA encoding eukaryotic initiation factor 2 α. Biochim. Biophys. Acta. 1991;1090:277–280. doi: 10.1016/0167-4781(91)90119-7. [DOI] [PubMed] [Google Scholar]
- Hardy S.J, Kurland C.G, Voynow P, Mora G. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- Kapp L.D, Lorsch J.R. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 2004;335:923–936. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- La Teana A, Gualerzi C.O, Brimacombe R. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA. 1995;1:772–782. [PMC free article] [PubMed] [Google Scholar]
- Lomakin I.B, Kolupaeva V.G, Marintchev A, Wagner G, Pestova T.V. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin I.B, Shirokikh N.E, Yusupov M.M, Hellen C.U.T, Pestova T.V. The fidelity of translation initiation: Reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Fekete C.A, Grczynski Z, Lorsch J.R. A conformational change in the eukaryotic translation preinitation complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Madjar J.J, Arpin M, Buisson M, Reboud J.P. Spot position of rat liver ribosomal proteins by four different two-dimensional electrophoreses in polyacrylamide gel. Mol. Gen. Genet. 1979;171:121–134. doi: 10.1007/BF00269998. [DOI] [PubMed] [Google Scholar]
- Nikiforov T.T, Connolly B.A. Oligodeoxynucleotides containing 4-thiothymidine and 6-thiodeoxyguanosine as affinity labels for the Eco RV restriction endonuclease and modification methylase. Nucleic Acids Res. 1992;20:1209–1214. doi: 10.1093/nar/20.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J.M, Brodersen D.E, Clemons W.M, Jr., Tarry M.J, Carter A.P, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Pestova T.V, Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V, Hellen C.U.T, Shatsky I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V, Borukhov S.I, Hellen C.U.T. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova T.V, Lomakin I.B, Lee J.H, Choi S.K, Dever T.E, Hellen C.U.T. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Rinke-Appel J, Junke N, Brimacombe R, Dukudovskaya S, Dontsova O, Bogdanov A. Site-directed cross-linking of mRNA analogues to 16S ribosomal RNA; A complete scan of cross-links from all positions between ‘+1’ and ‘+16’ on the mRNA, downstream from the decoding site. Nucleic Acids Res. 1993;21:2853–2859. doi: 10.1093/nar/21.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh S, Hijikata M, Handa H, Shimotohno K. Caspase-mediated cleavage of eukaryotic translation initiation factor subunit 2α. Biochem. J. 1999;342:65–70. [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jägow G. Tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sergiev P.V, Lavrik I.N, Wlasoff V.A, Dokudovskaya S.S, Dontsova O.A, Bogdanov A.A, Brimacombe R. The path of mRNA through the bacterial ribosome: A site-directed crosslinking study using new photoreactive derivatives of guanosine and uridine. RNA. 1997;3:464–475. [PMC free article] [PubMed] [Google Scholar]
- Shapkina T.G, Dolan M.A, Babin P, Wollenzien P. Initiation factor 3-induced structural changes in the 30S ribosomal subunit and in complexes containing tRNAfMet and mRNA. J. Mol. Biol. 2000;299:615–628. doi: 10.1006/jmbi.2000.3774. [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov S.I, Hellen C.U.T, Pestova T.V. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes & Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Nielsen K.H, Zhang F, Fekete C.A, Hinnebusch A.G. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly B.T, Brodersen D.E, Clemons W.M, Jr., Morgan-Warren R.J, Carter A.P, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Yatime L, Schmitt E, Blanquet S, Mechulam Y. Functional molecular mapping of archaeal translation initiation factor 2. J. Biol. Chem. 2004;279:15984–15993. doi: 10.1074/jbc.M311561200. [DOI] [PubMed] [Google Scholar]
- Yoon H, Donahue T.F. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol. Cell. Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov M.M, Spirin A.S. Hot tritium bombardment technique for ribosome surface topography. Methods Enzymol. 1988;164:426–439. doi: 10.1016/s0076-6879(88)64059-6. [DOI] [PubMed] [Google Scholar]
- Yusupova G.Z, Yusupov M.M, Cate J.H, Noller H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]