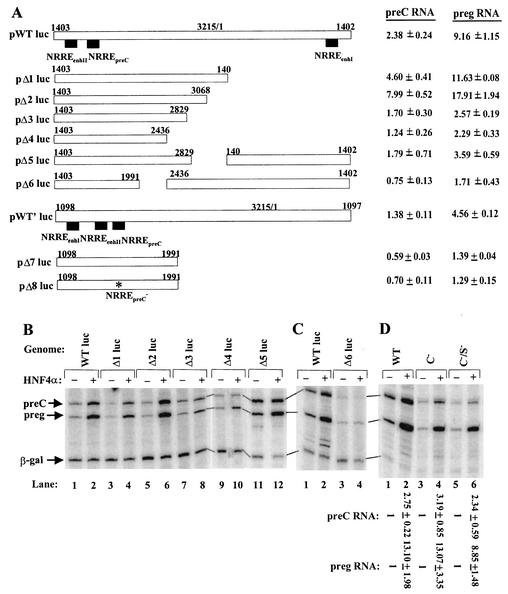
FIG. 8.
Mapping of regions in the HBV genome essential for transcriptional activation by HNF4α. (A) Schematic diagrams of the deletion variants of pWTluc and pWT′luc studied here. Plasmid pWT′luc is identical to pWTluc except for the permutation of the end points of the full-length HBV genome inserted into pGL2. Solid rectangles, locations of the three NRREs in pWTluc and pWT′luc; ∗ (in pΔ8luc), base substitutions in nt 1759, 1765, and 1768 in the NRREpreC− mutant previously described (48). preg, pregenomic. (B and C) Effects of deletions on activation of pregenomic RNA synthesis by HNF4α. WT, wild type. (D) Lack of effect of absence of C and S proteins on activation of synthesis of pregenomic RNA by HNF4α. Autoradiograms show the results of primer extension assays used to quantify the pre-C and pregenomic RNAs synthesized in Huh7 cells cotransfected with the indicated plasmids. The sequences of plasmids pC− and pC−/S− are described in Materials and Methods. Numbers in panels A and D indicate the amounts of the viral RNAs synthesized from the indicated HBV plasmid when HNF4α was overexpressed relative to the amounts synthesized from the same HBV plasmid when it was cotransfected in parallel with the parental expression plasmid. These data are means ± standard errors of the data obtained from three experiments similar to the one for which results are shown here and in Fig. 9.