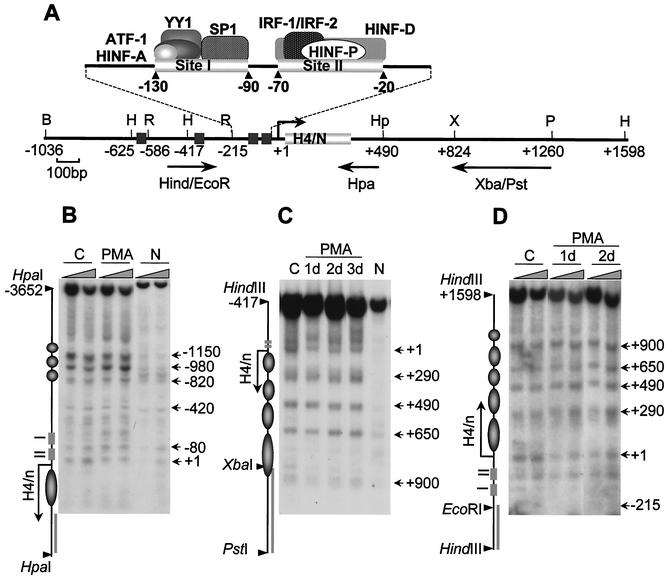
FIG. 2.
Maintenance of nucleosomal organization at the H4/n gene locus during HL-60 differentiation. (A) Restriction map of the human histone H4/n gene locus and functional organization of the proximal promoter. Arrows show DNA probes used for indirect end labeling to map nuclease accessibility of the H4/n locus. (B to D) Nuclei isolated from control (C) and PMA-treated (PMA) HL-60 cells, as well as naked DNA (N), were incubated with increasing concentrations of MNase for 5 min at room temperature. Purified DNA was digested with HpaI (B), PstI/HindIII (C), and HindIII (D). Southern blots were hybridized to radioactively labeled DNA fragments as follows: Hpa PCR probe (B), XbaI/PstI (C), and HindIII/EcoRI (D). The diagrams at the left of panels B, C, and D indicate the locations of transcription regulatory elements (rectangles), the positions of putative nucleosomes (circles and ovals), and the probes used for hybridization (solid bars). The arrows indicate the transcription start site and length of the protein-coding region. Numbers at the right indicate positions of MNase-sensitive sites.