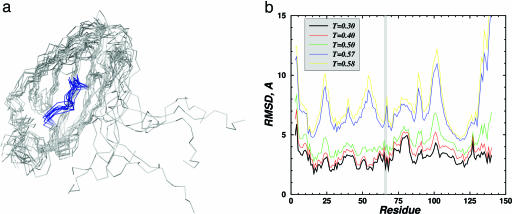
Fig. 1.
Structure of the large EGFP fragment (1–158 N-terminal amino acids) analyzed by DMD simulations. (a) Backbone representation of 10 folded and aligned structures of the large EGFP fragment obtained in DMD simulations at T = 0.3 (T is measured in ε/KB units). The segment from 62 to 70 amino acids, containing the chromophore-forming amino acids (T66, Y67, and G68), is colored blue. The C terminus of this polypeptide is very flexible because of a small number of contacts with the rest of the molecule, so the alignment was made by omitting these amino acids. (b) The root-mean-square deviation (RMSD) of each residue in the folded large EGFP fragment relative to the intact EGFP structure as a function of temperature. The chromophore-forming residues are in the shaded region, and their spatial arrangement at lower temperatures is essentially fixed, with deviation ≤2Å.