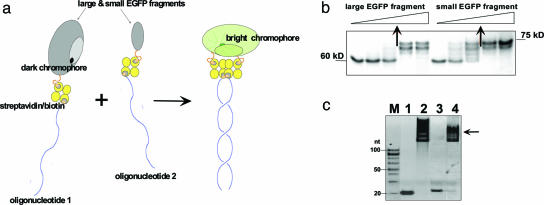
Fig. 3.
Design and assembly of a nucleic acid-supported EGFP complementation system with rapid signal response. (a) Schematics of the complementation of split fluorescent protein by DNA hybridization. Fluorescent protein (EGFP) is dissected into two nonfluorescent fragments, one of which contains preformed chromophore capable of bright fluorescence within a full-size protein. Both protein fragments are linked to complementary oligonucleotides via biotin–streptavidin interactions. In our protocol, we endeavor to create a 1:1:1 ratio of protein/streptavidin/oligonucleotide complex. In a mixture, the two nucleoprotein constructs associate by sequence-specific duplex DNA formation, which triggers complementation of the large and small EGFP fragments, resulting in fast development of fluorescence. (b) Gel-shift assay (10% SDS/PAGE) showing binding of increased amounts of biotinylated EGFP fragments with a fixed amount of streptavidin (2 μg; 60-kDa band). Arrows indicate the protein amounts resulting in 1:1 complexes (70- to 75-kDa bands), which correspond to ≥70% yield of biotinylation. (c) Gel-shift assay (10% PAGE) demonstrating the formation of 1:1:1 tripartite molecular constructions depicted in Fig. 1a and comprising the large or small EGFP fragment, streptavidin, and a corresponding oligonucleotide (see Table 1, which is published as supporting information on the PNAS web site, for their sequences). Lanes 1 and 2, biotinylated oligo 1 in the absence (1) or presence (2) of the large EGFP fragment coupled to streptavidin; lanes 3 and 4, biotinylated oligonucleotide 2 in the absence (3) or presence (4) of the small EGFP fragment coupled to streptavidin; M, 20-bp size marker. Arrow marks the position of the required oligonucleotide–protein complexes that are strongly shifted upward as expected.