Abstract
The deep subseafloor biosphere is among the least-understood habitats on Earth, even though the huge microbial biomass therein plays an important role for potential long-term controls on global biogeochemical cycles. We report here the vertical and geographical distribution of microbes and their phylogenetic diversities in deeply buried marine sediments of the Pacific Ocean Margins. During the Ocean Drilling Program Legs 201 and 204, we obtained sediment cores from the Peru and Cascadia Margins that varied with respect to the presence of dissolved methane and methane hydrate. To examine differences in prokaryotic distribution patterns in sediments with or without methane hydrates, we studied >2,800 clones possessing partial sequences (400–500 bp) of the 16S rRNA gene and 348 representative clone sequences (≈1 kbp) from the two geographically separated subseafloor environments. Archaea of the uncultivated Deep-Sea Archaeal Group were consistently the dominant phylotype in sediments associated with methane hydrate. Sediment cores lacking methane hydrates displayed few or no Deep-Sea Archaeal Group phylotypes. Bacterial communities in the methane hydrate-bearing sediments were dominated by members of the JS1 group, Planctomycetes, and Chloroflexi. Results from cluster and principal component analyses, which include previously reported data from the West and East Pacific Margins, suggest that, for these locations in the Pacific Ocean, prokaryotic communities from methane hydrate-bearing sediment cores are distinct from those in hydrate-free cores. The recognition of which microbial groups prevail under distinctive subseafloor environments is a significant step toward determining the role these communities play in Earth’s essential biogeochemical processes.
Keywords: 16S rRNA gene, deep biosphere, microbial diversity
Prokaryotic biomass in deep marine sediments exceeds 105 microbial cells/cm3 even at depths close to 1,000 m below the seafloor (mbsf) (1, 2). Extrapolation of these numbers to a global scale indicates that these deeply buried cells may represent one-tenth to one-third of living biomass on Earth (3). Despite the vast contribution of living biomass, relationships between the microbial community structure and distribution and the geophysical and geological conditions in subseafloor environments remain largely unknown.
Of particular interest in this regard are the microbial communities that contribute to the vast stores of dissolved and hydrated methane in deep marine sediments. Approximately 500–2,500 gigatonnes of total methane carbon are stored as gas hydrate or free gas in continental margins (4) and most of this methane is produced biologically (5). Recent investigations of sediments collected along the Nankai Trough reveal that diverse microbial communities are associated with methane hydrate-bearing sediments (6–8). One such study also resulted in the isolation of Methanoculleus submarinus, a new species of methanogen (9). In sediments of the Sea of Okhotsk, where the presence of methane hydrate is inferred by seismic surveys (10), microbial communities in pelagic clay layers are similar to those in hydrate-bearing sediments at the Nankai Trough (6), whereas ones in volcanic ash layers are clearly different (11). On the Cascadia Margin in the eastern Pacific Ocean, where only a few deep sediment samples from methane hydrate zones have been examined, microbial communities bear little similarity to those in western Pacific sediments (12, 13). It hence remains unknown whether the presence of methane hydrate affects microbial communities in the deep subseafloor biosphere.
To examine the potential significance of hydrates in determining biogeographical distribution and phylogenetic diversity of deeply buried prokaryotes, we extensively surveyed archaeal and bacterial communities down through six boreholes cored by the Ocean Drilling Program (ODP). The sample sites were located in three general areas and included offshore South and North America (Peru and Cascadia Margins) and the Eastern Equatorial Pacific (Fig. 4 and Table 1, which are published as supporting information on the PNAS web site). We here report that diverse microbial communities in subseafloor sediments on the Pacific Ocean Margin can be statistically distinguished based on the presence or absence of methane hydrate.
Results
Cell Abundance and Quantification of Archaeal 16S rRNA Genes.
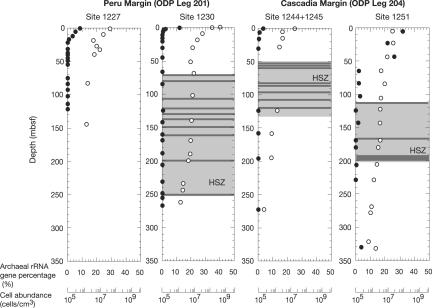
Microbial abundances estimated by acridine orange direct counts were between 106 and 107 cells/cm3 throughout most of the sediment columns at the Peru Margin and Cascadia Margin sites (Fig. 1) (14). Quantitative real-time PCR (Q-PCR) analysis based on 16S rRNA genes revealed that prokaryotic communities in deep marine sediments are mainly composed of Bacteria. Down through all of the sites drilled, Archaea reached the highest proportion of the prokaryotic community near the seafloor and decreased in proportion with depth (Fig. 1). The highest relative abundance of Archaea was observed at Site 1251, where Archaea comprised 30% of the total prokaryotic community near the seafloor. At all sites, the percentage of archaeal 16S rRNA genes obtained was very small (<0.01% of total prokaryotic signature) when the hydrate stability zone was reached.
Fig. 1.
Relative abundances of archaeal 16S rRNA genes (•) and total prokaryotic cells (○) in sediment cores from the Peru and Cascadia Margins. Abundances of archaeal genes were evaluated by Q-PCR of 16S rRNA genes by using domain specific primers and probe sets. Total prokaryotic cell numbers were calculated by fluorescent microscopic counts of cells stained by acridine orange. The depth range of the hydrate stability zone (HSZ) is indicated by the light gray box. The locations of hydrate as confirmed by low temperature anomalies in the samples and by shipboard observations are indicated by gray lines.
Archaeal Community Structure.
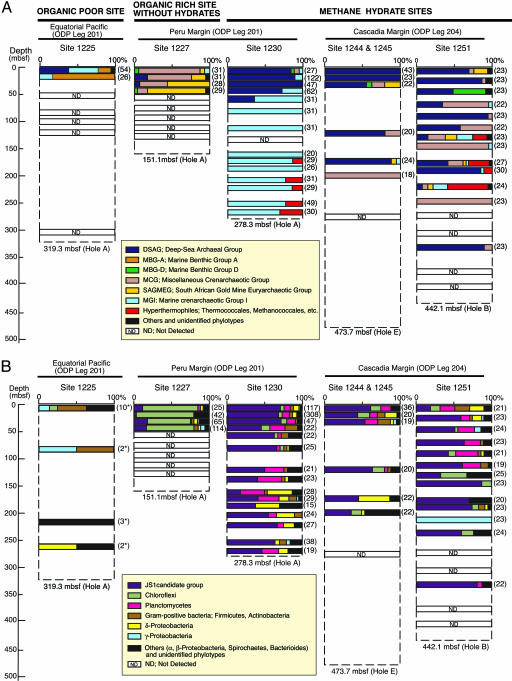
Archaeal diversity was assessed by phylogenetic analyses of 1,476 archaeal 16S rRNA gene sequences. The results showed that remarkable differences in the archaeal community structure were found depending on whether samples had been acquired from methane hydrate-bearing sites or hydrate-free sites. Archaeal communities at the hydrate-free, organic-poor Site 1225 in the Eastern Equatorial Pacific were dominated by Marine Crenarchaeotic Group I (MGI) and Marine Benthic Group A (Fig. 2A), both uncultivated groups that are widely distributed in seawater and shallow marine sediments (e.g., refs.15 and 16). At the hydrate-free organic-rich Site 1227 on the Peru Margin, Miscellaneous Crenarchaeotic Group (MCG), and South African Gold Mine Euryarchaeotic Group (SAGMEG) were the predominant Archaea detected (Fig. 2A). MCG has been detected in several terrestrial and marine environments (e.g., refs. 11,17, and 18), whereas SAGMEG phylotypes were originally reported from an ultradeep South African Gold Mine (18).
Fig. 2.
Phylogenetic community structures based on 16S rRNA gene clone libraries of domains Archaea (A) and Bacteria (B) from ODP sediment core samples. Numbers of clones examined at each depth are indicated in parentheses. The phylogenetic affiliation of each clone sequence was determined by similarity analysis of 400–500 bp of 16S rRNA gene sequences. In each column diagram, the relative abundances of clones classified with the (sub)phylum level are shown. Bacterial community structures at Site 1225 were evaluated by sequencing 200 bp of PCR fragments obtained by denaturing gradient gel electrophoresis (DGGE) analysis, and the number of major bands is indicated in parentheses with asterisk. Phylogenetic trees involving ≈1 kbp sequences of 348 representative phylotypes are shown in Fig. 5.
In marked contrast, clone libraries from hydrate-bearing sites of the Cascadia and Peru Margins were dominated by the Deep-Sea Archaeal Group (DSAG; alternatively designated as Marine Benthic Group B) (11, 16, 17). DSAGs have previously been reported from deep sediments in hydrate zones at the Nankai Trough (6) and the Sea of Okhotsk (11) as predominant archaeal phylotypes. Although archaeal populations at Sites 1230 (169–267 mbsf) and 1251 (123–304 mbsf) were below the detection limit for reliable quantification (0.01% of total prokaryotes), we unexpectedly detected sequences related to Marine Crenarchaeotic Group I (MGI) and putative (hyper-)thermophiles (e.g., Methanocaldococcus, Mthanothermococcus, Pyrococcus, Thermococcus, and Archaeoglobus relatives) from deep sediments (Fig. 2A). We detected only two putative mesophilic methanogen clones, related to Methanosarcina acetivorans (85.9% similarity) and Methanoculleus palmolei (98.8% similarity), in sediment horizons at 22.7 and 43.2 mbsf at Site 1251 (see Fig. 5E, which is published as supporting information on the PNAS web site).
Detection of Methyl Coenzyme M Reductase α-Subunit (mcrA) Genes from Peru Margin Sites.
Methyl coenzyme M reductase is a key enzyme for the terminal step in methanogenesis. The mcrA gene was obtained only at a depth of 44.3 mbsf at Site 1230 by nested PCR. Phylogenetic analysis indicated similarity to the mcrA gene of Methanosaeta concilii; a methanogen that was not detectable by the 16S rRNA gene-based survey (Fig. 6, which is published as supporting information on the PNAS web site).
Bacterial Community Structure.
A total of 1,343 bacterial 16S rRNA sequences were analyzed. There was, again, a clear difference in bacterial community structure between hydrate-containing (Site 1230 and Cascadia cores) and hydrate-free (Site 1227 and Eastern Equatorial Pacific) sites (Fig. 3B). As for the archaeal 16S rRNA genes, bacterial rRNA genes from Site 1227 were amplified only from shallow sediments (<50 mbsf). Nearly 80% of clones were affiliated with Chloroflexi (Fig. 2B). The phylum Chloroflexi is classified into four major subphyla (I, II, III, and IV) (19). Phylogenetic analyses suggest that sequences from deep marine sediments were affiliated with the subphyla I, II, and IV and a new cluster consisting only of subseafloor sequences (Fig. 5A). These Chloroflexi were detected at low (5–10%) clonal frequencies in samples from hydrate-bearing sites of the Peru and Cascadia Margins (Fig. 2B).
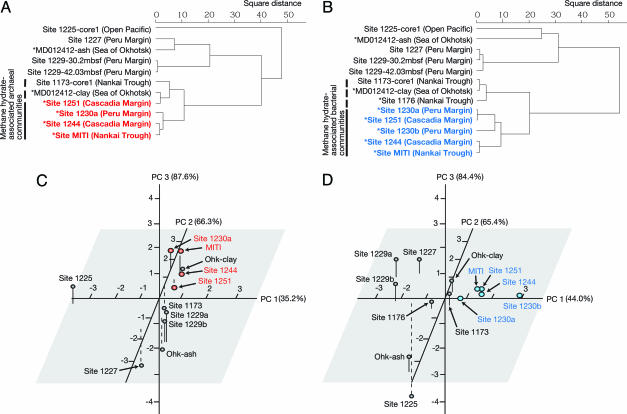
Fig. 3.
Statistical evaluation of microbial community structures based on clonal frequencies of 16S rRNA gene phylotypes from various marine sediments on the Pacific Ocean Margin. Cluster analysis of archaeal (A) and bacterial (B) communities. Scale bars indicate the square distance determined by the Ward method. The sampling sites, where hydrates are believed to exist nearby (based on seismic surveys), are marked by asterisks, and the sites where methane hydrates were directly observed in the cores are marked by bold colored letters. Shown is principal component (PC) analysis of archaeal (C) and bacterial (D) communities. The percent variance explained by each PC is shown in parentheses. Individual samples are designated in PC space according to the ODP sites from which they were acquired. Additional data points are shown for published clonal frequencies as noted in the text. The sites where methane hydrates were observed in the cores are marked by colored dots.
In the methane hydrate-bearing sediments, a total of 686 clone sequences (53.6% of the total bacterial clones) were affiliated with the uncultured JS1 group (20). Planctomycetes were also detected as predominant phylotypes (Fig. 2B). These phylotypes were not closely related to isolates such as Planctomyces and Pirellula. Within the δ-Proteobacteria, sequences related to genera Desulfobacterium, Syntrophus, Desulforhopalus, Pelobacter, Desulfococcus, and Desulfosarcina were detected. Small numbers of Actinobacteria, α-, β-, and γ-Proteobacteria, Bacteroidetes, Chlamydia, Defferribacteres, Firmicutes, Flavobacteria, Spirochaetes, candidate divisions OP1, OP3, OP8, OP10, OP11, WS1, and WS3, and several unclassified branches were found in deep sediments, accounting for a few percent of the total (Fig. 2B; phylogenetic trees in Fig. 5).
Discussion
Archaea and Bacteria Abundances.
Prokaryotic communities in deep marine sediments assessed by DNA appear to be composed mostly of Bacteria (Fig. 1), even though Q-PCR does not allow us to accurately quantify cell numbers because of differences in the number of gene copy per cell and of PCR biases. A high frequency of bacterial DNA has also been reported in deep sediments from the Sea of Okhotsk (11) and other sites on the Peru Margin (21, 22). The abundance of archaeal rRNA genes (0.01∼30%) was relatively high near the seafloor. This suggests that the upward flow of methane or hydrocarbons from the underlying hydrate stability zone and input of oxidants and organic nutrients from the overlying water column lead to highest archaeal populations in shallow sediments. At Site 1251, at the base of South Hydrate Ridge, archaeal gene abundances were higher (≈30%) and extended deeper into the sediments than at other sites. High sedimentation rates may lead to high availability of buried nutrients to greater depths than at sites, with lower rates where diagenesis leads to depletion of available nutrients in shallower sediment (23).
Biogeographical Distribution of Subseafloor Microbes.
Cluster analyses and principal component analyses indicate that microbial communities at methane hydrate sites are statistically similar regardless of location in the Pacific Ocean (Fig. 3). Bacterial communities at sites with hydrates (i.e., in Sites 1230, 1244, 1251, and MITI) formed a single clade, whereas communities in hydrate-free sites at the Peru Margin (Sites 1227 and 1229), Sea of Okhotsk (MD012412), and Nankai Trough (Sites 1173 and 1176) clustered in a different group (Fig. 3 B and D). Whereas seismic surveys indicated the presence of methane hydrates in areas close to Sites MD012412 and 1176 (10, 24), methane hydrate was not observed in these cores.
Members of the JS1 group were key bacterial representatives at methane hydrate sites. Clone library analyses suggested that the frequency of JS1 bacteria in hydrate sites was notably higher than in hydrate-free sites (often >50% of the representative clones for any given sample), suggesting that these bacteria may prefer sedimentary habitats with high concentrations of methane associated with hydrates. JS1 bacteria were uniform in distribution with depth at hydrate sites.
The phylum Chloroflexi is a dominant group at organic-rich sites lacking hydrates. The frequency of sequences related to Chloroflexi was higher in nonhydrate than in hydrate-bearing sediments. This was a key factor that distinguished hydrate sites from hydrate-free sites in cluster and principal component analyses (Fig. 3 B and D). The bacterial community at Site 1227, which is geographically close to the methane hydrate site (Site 1230), was mainly composed of Chloroflexi (Fig. 2B). The dominance of Chloroflexi sequences has also been reported for other organic-rich sediments without hydrates, such as Site 1229 on the Peru Margin (22). Our results indicate that sites may be spatially close but yet have distinct microbial communities. Habitat preference of subseafloor microbes may be affected by the presence of methane hydrate, the formation of which is controlled by a combination of geochemical (e.g., methane concentration, salinity) and geophysical (e.g., depth-related pressure, temperature, and hydrological flow) factors.
The archaeal community structure also appears to be affected by the presence of methane hydrate. Cluster and principal component analyses show that archaeal communities in sediments above hydrates are statistically differentiable from those in hydrate-free sediments at Sites 1225, 1227, and 1229 or in ash layers in the Sea of Okhotsk (Fig. 3 A and C). Sequence analyses indicated that archaeal communities in sediments above hydrates were commonly dominated by members of DSAG, irrespective of whether the samples came from the eastern or western Pacific. At hydrate-free sites (e.g., Sites 1227 and 1229), DSAG were also present, however as minority components microbial communities.
Controversially, MCG was the predominant archaeal component at hydrate-free Sites 1227 and 1229, the sediments of which contained a high concentration of methane (Fig. 2A) (14, 22). The predominance of MCG was also observed at several deeper horizons below the hydrate stability zone from the Cascadia Margin. In sediments from the Sea of Okhotsk, MCG was abundant in volcanic ash layers, whereas DSAG was the predominant archaeal component in pelagic clay layers (11). These observations suggest that microbial communities can be stratified in subseafloor sediments, and both lithological (e.g., pore space, grain size, mineral composition, sedimentation rate) and geochemical (e.g., pore water chemistry, methane concentration, and presence of hydrates) characteristics may affect microbial habitats in deep marine sediments.
Physiological Implications.
Although we cannot determine the physiologies of uncultivated subseafloor microbes, such as the JS1 bacteria, our observations suggest that members of the JS1 group preferentially inhabit strictly anaerobic organic-rich environments associated with hydrates. JS1 bacteria were uniformly distributed as the predominant bacterial representatives throughout the hydrate-bearing cores (Fig. 2B), even in sulfate-free deeper zones (14). This suggests that this group is probably not responsible for sulfate reduction. On the other hand, Q-PCR and clone library analyses indicated that the DSAG was generally abundant in the sulfate reduction zone in shallow sediments above hydrates where the upward methane diffusion from hydrates occurred (Figs. 1 and 2A). We infer that the uncultivated DSAG may play a role in biogeochemical processes such as sulfate reduction and methane oxidation (25).
In contrast to the JS1 group and DSAG, several species within the Chloroflexi have been isolated and identified. The 16S rRNA gene sequences of Chloroflexi from deep marine sediments were affiliated with the subphyla I, II, and IV and a new subphylum and clearly differed from the subphylum III representing phototrophs or aerobic heterotrophs. Although there is no isolate within the subphylum IV, two genera Anaerolinea and Caldilinea within the subphylum I were recently characterized (26). Both species can grow anaerobically and chemoorganotrophically on a number of carbohydrates and amino acids in the presence of yeast extract (26). Interestingly, the growth of Anaerolinea and its relatives can be stimulated in cocultures with hydrogenotrophic methanogen (Y. Kamagata, personal communication; see ref. 26). Within the subphylum II, sequences from deep marine sediments were most closely related to the genus Dehalococcoides. This microorganism is a facultatively hydrogenotrophic and heterotrophic anaerobic bacterium that can use chloride compounds as electron acceptors (27). Given these physiological characteristics, it is possible that closely related members of heterotrophic Chloroflexi living in deep marine sediments grow syntrophically with hydrogenotrophic Chloroflexi species or other hydrogenotrophic microbes such as methanogens.
Proteobacterial clone sequences were dominant bacterial components from several horizons. At Site 1251, 204.2 mbsf, all sequences in the clone library clustered with the same phylotype and the representative sequence showed 96.1% similarity with Marinobacter aquaeolei (Figs. 2B and 5B). In a previous study of the Sea of Okhotsk, α- and γ-Proteobacteria dominated in volcanic ash layers, and the genera Halomonas, Marinobacter, and Sulfitobacter were frequently isolated (11). At Site 1230, members of the genera Halomonas, Marinobacter, Shewanella, Photobacterium, and Vibrio were cultivated (14, 28). Most cultivable members in deep marine sediments were found to be facultatively anaerobic, heterotrophic, piezophilic (tolerant or adapted to high pressure), or halophilic bacteria (11, 14, 28, 29). Anaerobic and heterotrophic respiration or fermentation of Proteobacteria probably contributes to the degradation of organic substrates in deeply buried marine sediments.
Microbial Communities at Sulfate–Methane Transition Zones.
In sulfate–methane transition zones at shallow methane seeps, anaerobic methane-oxidizing archaea (ANME) are believed to be frequently responsible for anaerobic oxidation of methane (AOM) coupled to sulfate reduction by δ-Proteobacteria (e.g., refs. 30–32). We did not detect ANME sequences in the examined sediments; however, sequences of sulfate reducers within the Desulfococcus/Desulfosarcina group, which is a typical cluster of δ-Proteobacteria associated with AOM, were detected from several horizons (Fig. 5C). At the sulfate methane transition zones of Sites 1227 (near 50 mbsf) and 1230 (near 9 mbsf) (14, 25), archaeal communities were mainly composed of SAGMEG and DSAG, respectively (Fig. 2A). At Site 1229, the archaeal community was mainly composed of MCG (22, 25). At the Sea of Okhotsk, two clones belonging to the ANME-1 cluster were detected in DSAG-dominated pelagic clay at 14.7 mbsf (OHKA3.36: GenBank accession no. AB094534) (11); however, geochemical data indicating sulfate and methane concentrations were elusive. Given these results, we assume that (i) ANME groups are absent from our sampling sites, (ii) the population of ANME is below the detection limit, (iii) ANME groups inhabit narrow (or patchy) horizons we have not sampled, or (iv) some other unidentified phylotypes are responsible for AOM.
Where Does Methane Come From?
We detected a few clones of 16S rRNA and mcrA genes of known mesophilic methanogens in methane hydrate-bearing sediments. Using PCR amplification with methanogen-specific primers, 16S rRNA or mcrA genes of methanogens such as Methanobacteriales have been retrieved from deep sediments at the Cascadia (13) and Peru Margins (Site 1230 in this study and ref. 22). Methanogens were not detected with universal primer sets in the Nankai Trough (6); however, Nankai Trough sediments collected at the same time yielded a methanogen isolate (9). These results suggest that, whereas methanogens are indeed present in hydrate-bearing sediments, their population size appears to be small. We also detected the sequences related to (hyper)thermophilic methanogens as minor archaeal populations, postulating that sequences are from deeply buried microbial relicts, referred to as the “Paleome” (33).
Given the scarcity of known methanogens in deep marine sediments rich in biologically produced methane and the successful detection of mcrA genes, several scenarios may explain the presence of biogenic methane: (i) production by unidentified prokaryotes and/or via an uncharacterized methanogenic pathway; (ii) production in situ by a numerically small population of mesophilic methanogens for geological timescale; and/or (iii) production elsewhere with methane supply via fluid flow from either vertically or laterally distinct terranes on the accretionary margins. One or more of these explanations may account for our observations.
Conclusion
Our data suggest that previously unidentified prokaryotic communities such as the JS1 and DSAG groups occur widely in organic-rich deep marine sediments associated with methane hydrates along the Pacific Ocean Margin. Microbial communities can be stratified in deep marine sediments, and surrounding geochemical and geological settings strongly affect the community structure. Almost all prokaryotes that we detected represent uncultivated and physiologically uncharacterized assemblages. Nevertheless, the recognition of microbial populations that consistently occur in the presence of methane hydrates serves as a starting point for defining their ecological and biogeochemical significances. We anticipate that future studies of the microbes in near-shore deep marine sediments will clarify their role in the formation of methane hydrates, determine how these cells affect the cycling of carbon in subsurface strata, and begin to establish the physiological properties that permit their survival at depth.
Materials and Methods
Sampling Site.
Site location, water depth, core length, presence of methane hydrate, and primary lithological characteristics of the cores are summarized in Fig. 4 and Table 1. Details of the site description are provided in Supporting Text, which is published as supporting information on the PNAS web site (also see refs. 14 and 23).
DNA Extraction and Purification.
DNA was extracted from 10 g of wet sediment, as described (11, 32). In a final cleanup step to remove PCR inhibitors, the extract was applied to the DNA IQ System (Promega). Because of very low recovery of DNA, a modified DNA extraction method, described previously as Protocol ENZ (34), was applied to the organic-poor sediments at Site 1225.
Quantitative PCR Analysis of Archaeal and Bacterial 16S rRNA Genes.
The abundance of archaeal and total prokaryotic 16S rRNA genes was evaluated via Q-PCR with Archaea-specific and universal (Archaea and Bacteria) TaqMan probes and primer sets (35). Relative abundances of archaeal rRNA genes within total prokaryotic rRNA genes were calculated from yields of PCR-amplified fragments per sample.
Cloning and Sequencing of PCR-Amplified 16S rRNA and mcrA Genes.
Microbial 16S rRNA and mcrA genes were PCR-amplified by using LA Taq polymerase with GC buffer I (Takara Bio, Tokyo). The conditions of all PCRs are provided in Supporting Text. PCR-amplified gene fragments were cloned and sequenced as described (11, 32).
Phylogenetic Analysis.
Based on ≈400–500 base pair sequences, similarities among all clones (>2,800 clones) were analyzed by using the fasta program equipped with dnasis software (Hitachi Software, Tokyo). The sequences having >97% (Archaea) and >95% (Bacteria) similarity were tentatively assigned to the same group or phylotype. A total of 348 representative clones were selected and sequenced from both strands. The 2% cutoff discrepancy between Archaea and Bacteria did not influence phylum level classification, as shown in Fig. 2. Phylogenetic trees were constructed by neighbor-joining analyses. Details are provided in Supporting Text.
Statistical Analysis.
To evaluate the similarity of microbial communities among sampling sites, compiled clonal frequencies of 16S rRNA genes within (sub)phylum levels were subjected to cluster analyses and principal component analyses by using the Web-based software black box (S. Aoki, Gunma University, Gunma, Japan). The following published clonal frequency data were included: sediment above hydrates at Site MITI in the Nankai Trough (6), pelagic clay and ash layer samples of MD012412 core in the Sea of Okhotsk (11), four deep sediment samples (1.2, 51.1, 98.5, and 194.0 mbsf) of ODP Site 1176 (7), a sediment sample at 4.1 mbsf from ODP Site 1173 (Leg 190) (8) in the Nankai Trough, and two sediment samples (30.2 and 42.0 mbsf) from ODP Site 1229 (Leg 201) (22) at the Peru Margin. The clone sequences were recategorized at the (sub)phylum level by phylogenetic analysis, as described above. For the community at Site 1230, the clone library data from sediments above and below 100 m were compiled as designated Sites 1230a and 1230b, respectively. For the archaeal community at Site 1251, the data from sediments above hydrates (≈100 m), where the significant archaeal abundance was observed by Q-PCR analysis, were evaluated. The progressions were constructed by percentage scores of clonal frequency, and 12 units of the major phyla were used in the analyses as follows: DSAG [Marine Benthic Group (MBG)-B], MGI, MCG, MBG-A, MBG-D, and SAGMEG for archaeal communities; JS1 group, Chloroflexi, Planctomyces, Firmicutes, Actinobacteria, and γ- and δ-Proteobacteria for bacterial communities.
Supplementary Material
Acknowledgments
We acknowledge the personnel of Legs 201 and 204 shipboard scientific parties for sampling ODP core sediments. We are grateful to K.-U. Hinrichs, C. House, and A. Schippers for useful discussions and to B. A. Cragg and R. J. Parkes for total cell counts in Peru Margin sediments during Leg 201. This study was supported by Integrative Graduate Education and Research Traineeships Grant DGE-9972759 from the National Aeronautics and Space Administration Astrobiology Institute (Subsurface Biosphere and Environmental Genomes). Support to F.S.C. and M.D. was provided by the U.S. Department of Energy, Office of Fossil Energy. Support to F.I. was partially provided by a Humboldt Foundation Research Fellowship. ODP is sponsored by the U.S. National Science Foundation and participating countries under the management of Joint Oceanographic Institutions, Inc.
Abbreviations
- ODP
Ocean Drilling Program
- Q-PCR
quantitative real-time PCR
- mbsf
meters below the seafloor
- MCG
Miscellaneous Crenarchaeotic Group
- DSAG
Deep-Sea Archaeal Group
Footnotes
References
- 1.Parkes R. J., Cragg B. A., Wellsbury P. Hydrogeol. J. 2000;8:11–28. [Google Scholar]
- 2.Parkes R. J., Cragg B. A., Bale S. J., Getliff J. M., Goodman K., Rochele P. A., Fry J. C., Weightman A. J., Harvey S. M. Nature. 1994;371:410–413. [Google Scholar]
- 3.Whitman W. B., Coleman D. C., Wiebe W. J. Proc. Natl. Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milkov A. V. Earth–Science Rev. 2004;66:183–197. [Google Scholar]
- 5.Kvenvolden K. A. Org. Geochem. 1995;23:997–1008. [Google Scholar]
- 6.Reed D. W., Fujita Y., Delwiche M. E., Blackwelder D. B., Sheridan P. P., Uchida T., Colwell F. S. Appl. Environ. Microbiol. 2002;68:3759–3770. doi: 10.1128/AEM.68.8.3759-3770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kormas K. A., Smith D. C., Edgcomb V. E., Teske A. FEMS Microbiol. Ecol. 2003;45:115–125. doi: 10.1016/S0168-6496(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 8.Newberry C. J., Webster G., Cragg B. A., Parkes R. J., Weightman A. J., Fry J. C. Environ. Microbiol. 2004;6:274–287. doi: 10.1111/j.1462-2920.2004.00568.x. [DOI] [PubMed] [Google Scholar]
- 9.Mikucki J. A., Liu Y., Delwiche M., Colwell F. S., Boone D. R. Appl. Environ. Microbiol. 2003;69:3311–3316. doi: 10.1128/AEM.69.6.3311-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamaki K., Nishimura K., Honza E. Cruise Report No. 11. Tokyo: Geological Survey of Japan; 1978. pp. 42–43. [Google Scholar]
- 11.Inagaki F., Suzuki M., Takai K., Oida H., Sakamoto T., Aoki K., Nealson K. H., Horikoshi K. Appl. Environ. Microbiol. 2003;69:7224–7235. doi: 10.1128/AEM.69.12.7224-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bidle K. A., Kastner M., Bartlett D. H. FEMS Microbiol. Lett. 1999;177:101–108. doi: 10.1111/j.1574-6968.1999.tb13719.x. [DOI] [PubMed] [Google Scholar]
- 13.Marchesi J. R., Weightman A. J., Cragg B. A., Parkes R. J., Fry J. C. FEMS Microbiol. Ecol. 2001;34:221–228. doi: 10.1111/j.1574-6941.2001.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 14.D’Hondt S., Jørgensen B. B., Miller D. J., Batzke A., Blake R., Cragg B. A., Cypionka H., Dickens G. R., Ferdelman T., Hinrichs K.-U., et al. Science. 2004;306:2216–2221. doi: 10.1126/science.1101155. [DOI] [PubMed] [Google Scholar]
- 15.Karner M. B., DeLong E. F., Karl D. M. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 16.Vetriani C., Jannasch H. W., MacGregor B. J., Stahl D. A., Reysenbach A.-L. Appl. Environ. Microbiol. 1999;65:4375–4384. doi: 10.1128/aem.65.10.4375-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takai K., Horikoshi K. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takai K., Moser D. P., DeFlaun M., Onstott T. C., Fredrickson J. K. Appl. Environ. Microbiol. 2001;67:5750–5760. doi: 10.1128/AEM.67.21.5750-5760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hugenholtz P., Goebel B. M., Pace N. R. J. Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster G., Parkes R. J., Fry J. C., Weightman A. J. Appl. Environ. Microbiol. 2004;70:5708–5713. doi: 10.1128/AEM.70.9.5708-5713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schippers A., Neretin L. N., Kallmeyer J., Ferdelman T., Cragg B. A., Parkes R. J., Jørgensen B. B. Nature. 2005;433:861–864. doi: 10.1038/nature03302. [DOI] [PubMed] [Google Scholar]
- 22.Parkes R. J., Webster G., Cragg B. A., Weightman A. J., Newberry C. J., Ferdelman T. G., Kallmeyer J., Jørgensen B. B., Aiello I. W., Fry J. C. Nature. 2005;436:390–394. doi: 10.1038/nature03796. [DOI] [PubMed] [Google Scholar]
- 23.Tréhu A. M., Bohrmann G., Rack F. R., Shipboard Scientific Party Proc. ODP Int. Rep; 2003. www-odp.tamu.edu/publications/204_IR/204ir.htm. [Google Scholar]
- 24.Baba K., Yamada Y. Resource Geol. 2004;54:11–24. [Google Scholar]
- 25.Biddle J. F., Lipp J. S., Lever M., Lloyd K., Sørensen K., Anderson R., Fredricks H. F., Evert M., Kelly T. J., Schrag D. P., et al. Proc. Natl. Acad. Sci. USA. 2006 in press. [Google Scholar]
- 26.Sekiguchi Y., Yamada T., Hanada S., Ohashi A., Harada H., Kamagata Y. Int. J. Syst. Evol. Microbiol. 2003;53:1843–1851. doi: 10.1099/ijs.0.02699-0. [DOI] [PubMed] [Google Scholar]
- 27.Maymo-Gatell X., Chien Y.-T., Gossett J. M., Zinder S. H. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 28.Biddle J. F., House C. H., Brenchley J. E. Proc. ODP Sci. Results; 2005. pp. 1–19. www-odp.tamu.edu/publications/201_SR/VOLUME/CHAPTERS/107.PDF. [Google Scholar]
- 29.Köpke B., Wilms R., Engelen B., Cypionka H., Sass H. Appl. Environ. Microbiol. 2005;71:7819–7830. doi: 10.1128/AEM.71.12.7819-7830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinrichs K.-U., Hayes J. M., Sylva S. P., Brewer P. G., DeLong E. F. Nature. 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 31.Boetius A., Ravenschlag K., Schubert C. J., Rickert D., Widdel F., Gieseke A., Amann R., Jørgensen B. B., Witte U., Pfannkuche O. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 32.Inagaki F., Tsunogai U., Suzuki M., Kosaka A., Machiyama H., Takai K., Nunoura T., Nealson K. H., Horikoshi K. Appl. Environ. Microbiol. 2004;70:7445–7455. doi: 10.1128/AEM.70.12.7445-7455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inagaki F., Okada H., Tsapin A, I., Nealson K. H. Astrobiology. 2005;5:141–153. doi: 10.1089/ast.2005.5.141. [DOI] [PubMed] [Google Scholar]
- 34.Sørensen K. B., Lauer A., Teske A. Geobiology. 2004;2:151–161. [Google Scholar]
- 35.Takai K., Horikoshi K. Appl. Environ. Microbiol. 2000;66:5066–5072. doi: 10.1128/aem.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.