Abstract
The lipids of enveloped viruses play critical roles in viral morphogenesis and infectivity. They are derived from the host membranes from which virus budding occurs, but the precise lipid composition has not been determined for any virus. Employing mass spectrometry, this study provides a quantitative analysis of the lipid constituents of HIV and a comprehensive comparison with its host membranes. Both a substantial enrichment of the unusual sphingolipid dihydrosphingomyelin and a loss of viral infectivity upon inhibition of sphingolipid biosynthesis in host cells are reported, establishing a critical role for this lipid class in the HIV replication cycle. Intriguingly, the overall lipid composition of native HIV membranes resembles detergent-resistant membrane microdomains and is strikingly different from that of host cell membranes. With this composition, the HIV lipidome provides strong evidence for the existence of lipid rafts in living cells.
Keywords: dihydrosphingomyelin, nano-electrospray ionization tandem mass spectrometry, lipid analysis, viral membrane
The nucleic acid and protein constituents of many viruses have been identified, quantitated, and characterized in detail, whereas a comprehensive and quantitative analysis of the lipid composition, including molecular species of each lipid class, has not been reported for any virus. Enveloped viruses acquire their membrane by budding from a host cell membrane, but previous reports already indicated that viral lipids may differ from those of their respective budding membrane. It was suggested, therefore, that these viruses bud from membrane microdomains, and this hypothesis has gained momentum in recent years in the context of the lipid raft model (for review of lipid rafts, see ref. 1). Many viruses are now believed to bud from lipid rafts, with this assignment being based on the coclustering of viral structural proteins with putative raft markers, their partitioning into buoyant fractions after membrane extraction with cold detergent, and the sensitivity of virus release and/or infectivity to cholesterol extraction (2). The main method to operationally define raft lipids and proteins was extraction with cold Triton X-100. This methodology leads to artificial aggregation of raft components, however, and therefore, can not be used to define raft microdomains in natural cell membranes. The present concept of lipid rafts is that they are dynamic assemblies of sphingolipids, cholesterol, and raft proteins that associate and dissociate on a rapid time scale. These assemblies can be induced to coalesce from specific raft clusters usually by protein–protein interactions, and these assemblies are the platforms that are used in membrane trafficking, signaling, and virus budding (3, 4). Most of the evidence for the existence of lipid rafts relies on indirect methods, however, both in the analysis of cell and viral membranes (5). The in vivo existence of lipid rafts remains extremely controversial, because evidence for their presence in intact biological membranes is still not conclusive, despite that such evidence has been sought for more than a decade (6).
HIV type 1 (HIV-1) is an enveloped retrovirus, which buds primarily from the plasma membrane of infected T cells. HIV-1 morphogenesis is driven by the viral Gag polyprotein, which has been shown to partially localize to detergent-resistant membranes (DRMs) in infected cells. This result and the observation that HIV-1 particles contain putative raft proteins led to the suggestion that HIV-1 budding occurs from lipid rafts (ref. 2 and references therein). Earlier support for this hypothesis came from a comparative analysis of lipid groups from HIV-1 particles and plasma membranes of producer cells that revealed an enrichment of cholesterol and sphingomyelin (SM) in the virus (7, 8). Furthermore, cholesterol depletion impaired HIV-1 release and infectivity (9). A cholesterol requirement for virus morphogenesis has also been reported for alphaviruses (10), which are suggested not to bud from lipid rafts (11), and the association of HIV-1 budding and lipid rafts remains controversial.
Results and Discussion
Quantitative Lipid Analysis of HIV-1.
To investigate HIV-1 envelopment, we performed a quantitative composition analysis of viral lipids, including analysis of molecular species. HIV-1 particles were purified from the medium of the infected T cell line, MT-4, by velocity gradient centrifugation. This protocol yields essentially vesicle-free virus preparations (12). Lipid extracts from HIV-1 and from uninfected or infected MT-4 cells were subjected to nano-electrospray ionization tandem mass spectrometry (13). As control for purity, tissue culture supernatant from uninfected cells was subjected to the same procedure, and no lipid background signal was detectable in this case (data not shown). No significant differences in lipid composition were observed between infected and uninfected cells.
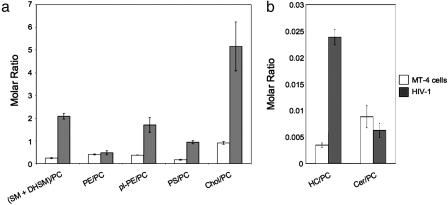
Table 1 shows that the phospholipid composition of HIV-1 differed significantly from that of MT-4 cells. Because of technical limitations in the homogenous preparation of plasma membranes, the lipid composition was analyzed for total cellular membranes. Phosphatidylcholine (PC) and phosphatidylethanolamine (PE), as major phospholipids of mammalian membranes, were reduced in viral membranes by a factor of 2.7 and 2.1, respectively. In contrast, SM and dihydrosphingomyelin (DHSM) (collectively referred to as SMs), 1-alkenyl,2-acylglycerophosphoethanolamine [referred to as plasmalogen PE (pl-PE, plasmenylethanolamine)], and phosphatidylserine (PS) were enriched in viral membranes by a factor of 3.2 (SMs), 2.1 (PS), and 1.7 (pl-PE) (for structures of lipids analyzed, see Fig. 6, which is published as supporting information on the PNAS web site). Cholesterol was also increased strongly in viral membranes: The ratio of cholesterol to total phospholipids was 0.39 in host and 0.83 in viral membranes (Fig. 1a). Likewise, monohexosylceramide (HC) was increased by a factor of 2.6 from 0.0015 mol % (host membranes) to 0.0038 mol % (virus). Employing separation by thin layer chromatography of glycolipids extracted from MT-4 cells, the HC species was identified as glucosylceramide (data not shown). Ceramide (Cer) was reduced by a factor of 3.8 in viral membranes (0.004 mol % in host and 0.001 mol % in viral membranes) (Fig. 1b). Lysobisphosphatidic acid (LBPA), a cone-shaped phospholipid, which has been shown to be required for vesicle formation at the late endosome (14), could not be quantified by mass spectrometry because it cannot be unequivocally distinguished from its mass isomer phosphatidylglycerol.
Table 1.
Phospholipid composition of MT-4 cells and HIV-1
| MT-4 cells(mol % ± SD) | HIV-1(mol % ± SD) | |
|---|---|---|
| PC | 43.0 ± 2.9 | 16.0 ± 1.0 |
| SM + DHSM | 10.4 ± 1.6 | 33.1 ± 1.2 |
| PE | 17.0 ± 1.5 | 8.2 ± 1.3 |
| pI-PE | 15.9 ± 0.5 | 27.0 ± 3.3 |
| PS | 7.4 ± 0.8 | 15.5 ± 2.2 |
Lipids were extracted and analyzed for phospholipid content as described in Materials and Methods. Values are expressed either as mol percentage of a given phospholipid to total phosphate (MT-4 cells) or as mol percentage of a given phospholipid to the total of all phospholipids quantified (HIV-1).
Fig. 1.
Lipid composition of MT-4 cells and HIV-1. Quantitative lipid analysis was performed as described in Materials and Methods. Data are displayed as molar ratio of individual lipid classes to PC. Error bars represent standard deviation of the mean.
An enrichment of sphingolipids and cholesterol is characteristic for DRM. A second hallmark of DRM is the enrichment of saturated lipid species such as dipalmitoyl-PC (15). Quantitative analysis of lipid subclasses revealed a 3.6-fold increase of saturated PC species in HIV-1 compared with the cellular membrane, resulting in a total of 40% saturated PC species in the HIV-1 membrane, with dipalmitoyl-PC alone representing ≈20% of total PC (see Fig. 7, which is published as supporting information on the PNAS web site). This enrichment was balanced by a reduction of diunsaturated and polyunsaturated species (2.5- and 2.3-fold, respectively), whereas monounsaturated species remained constant with a contribution of 36–38% (see Fig. 7b). The increase in saturated PC species is mainly attributed to short chain (up to 32 C atoms in acyl chains) PC species (see Fig. 7c), which were increased from 18% in cell membranes to 41% in viral membranes, at the expense of both medium (up to 36 C atoms) and long (≥38 C atoms) chain PC species. This tendency was also observed for PE, pl-PE, and PS, albeit less pronounced (see Figs. 8–10, which are published as supporting information on the PNAS web site). In contrast, no significant changes with regard to saturation or fatty acid chain length were observed for SM species (see Fig. 11, which is published as supporting information on the PNAS web site).
DHSM Is Highly Enriched in HIV-1 Membranes.
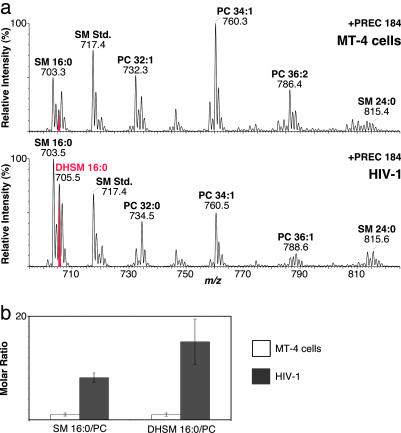
To determine the molecular species distribution of PC and SMs, we performed precursor ion scanning selecting for fragment ions of m/z 184 Da (+PREC 184). These ions are the most abundant fragment ions of PC and SM and correspond to the positively charged choline phosphate head group. Unexpectedly, we found a strong enrichment of the unusual sphingolipid DHSM in HIV-1 (Fig. 2). In contrast to SM, DHSM does not contain a 4,5-trans double bond in its sphinganine backbone. To validate the presence of DHSM, we subjected the ions of interest to collision-induced fragmentation and to mild basic treatment, thereby confirming their identity (for details see Fig. 12, which is published as supporting information on the PNAS web site). The major species of SM and DHSM in the viral and cellular membrane were SM d18:1/16:0 and DHSM d18:0/16:0, both containing an amide-linked palmitoyl group (Fig. 2a).
Fig. 2.
Enrichment of DHSM 16:0/PC in HIV-1. (a) Lipids of cells (Upper) and virus (Lower) were subjected to mass spectrometer analysis. Precursor ion scanning was performed by selecting for fragment ions of m/z 184 Da. Major peaks are labeled giving lipid class, number of total C atoms, and double bonds in acylated fatty acids. Spectra are normalized to the highest peak in the displayed mass range. (b) Quantitative analysis of SM and DHSM was performed as described in Materials and Methods. Data are displayed as molar ratios of the indicated lipid species to PC. Ratios determined for host cell membranes were set to 1. Error bars represent standard deviation of the mean.
Quantitation of SM and DHSM species was performed by employing the sensitive +PREC 184 scan (Fig. 2). There was no difference in the relative abundance of choline phosphate fragment ions generated from lipids containing either a sphingosine or a sphinganine backbone (Fig. 12d and ref. 16), and nonnatural SM species could therefore be used for quantitation of both lipids. As shown in Fig. 2b, the major species of both SM and DHSM were highly enriched in viral compared with cellular membranes: The molar ratio of SM 16:0/PC was increased almost 8-fold in HIV-1 (0.12 versus 0.95), and the molar ratio of DHSM 16:0/PC was increased by a factor of almost 15 (0.028 versus 0.41). Thus, the dihydro species of SM is enriched in HIV-1 even higher than SM, and the molecular species N-palmitoyl-DHSM alone constitutes ≈10% mol of total HIV-1 phospholipids. We also confirmed the presence of dihydroceramide as a potential precursor of DHSM as well as dihydromonohexosylceramide, but quantitation was not possible because of the low yield of the relevant fragment ions (16).
DHSM has not been reported as a component of HIV-1, which is not surprising given that data for other sphingolipid species in the HIV-1 membrane are also not available. So far, DHSM was described as a major constituent of biological membranes only in human lens extracts, where it accounts for 50% of all phospholipids (17). The function of DHSM is currently not known. DHSM in the cholesterol-rich membranes of the eye lens has been suggested to contribute to a greater resistance to oxidation and to the formation of cholesterol crystallites (18). Biophysical studies on liposomes comparing acyl-chain-matched SM and DHSM have shown that DHSM leads to formation of more ordered membranes with a higher melting temperature because of its less polar nature (18–20). Based on these results, DHSM was suggested to function as membrane organizer in laterally condensed liquid-ordered membrane domains, such as lipid rafts. Accordingly, the high amount of DHSM in the SM- and cholesterol-rich HIV-1 membrane may contribute to membrane order and physical stability of the extracellular virion but may also be important for resistance to oxidation. Conceivably, pl-PE, which is also enriched in the HIV-1 membrane, may contribute to this resistance as well, as plasmalogens have been discussed to function as antioxidants (for review, see ref. 21).
Inhibition of Sphingolipid Biosynthesis Reduces HIV-1 Infectivity.
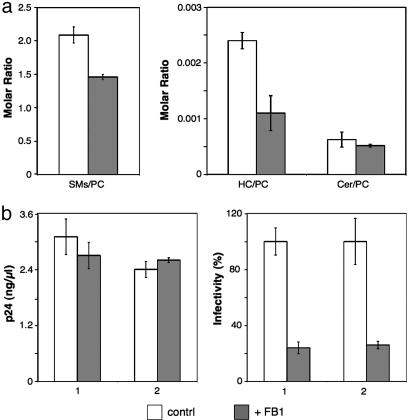
Previous studies have reported that inhibition of cholesterol biosynthesis in HIV-1-producing cells or cholesterol extraction from cell or virus membranes caused a decrease in virus production and infectivity (9, 22). To test whether sphingolipids are also crucial for HIV-1 formation and/or infectivity, we subjected infected MT-4 cells to fumonisin B1 (FB1) treatment. FB1 blocks both the de novo synthesis of sphingolipids (by inhibiting the synthesis of dihydroceramide, which serves as precursor to all sphingolipid species) as well as the salvage pathway (23). HIV-1 purified from FB1-treated cells exhibited a significantly reduced ratio of sphingolipids compared with PC, consistent with the expected effect of the inhibitor. Reductions of 19% (Cer), 30% (SMs), and 54% (HC), respectively, were observed for the different sphingolipids (Fig. 3a).
Fig. 3.
FB1 treatment of virus-producing cells reduces HIV-1 infectivity. HIV-1 infected MT-4 cells were cultured in the absence or presence of 50 μM FB1. (a) Quantitation of lipids from purified virus was performed by nano-electrospray ionization tandem mass spectrometry. Values are expressed as molar ratios of individual lipid classes to PC. (b) Quantitation of HIV-1 release (measured by antigen ELISA; Left) and infectivity of cell-free virus (measured by infection of TZM cells; untreated samples were set to 100%; Right). Bars 1 and 2 correspond to two independent infections, each performed in triplicate. Error bars represent standard deviation of the mean.
This reduction of sphingolipids (which was also observed in cellular membranes; data not shown) had no significant effect on virus release (Fig. 3b). A strong effect on HIV-1 infectivity was observed, however. HIV-1 released from FB1-treated cells was reproducibly 4-fold less infectious than virus from untreated cells (Fig. 3b). Accordingly, the concentration of sphingolipids in the HIV-1 membrane appears to be important for maintaining viral infectivity, although not being required for virus budding. Our results indicate that changes in the lipid composition of biological membranes may only be tolerable within a narrow range, because partial reduction of viral sphingolipids yielded a substantial decrease in HIV-1 infectivity. The conclusion that subtle changes in lipid composition can lead to significant functional alterations had been drawn previously from studies on transport processes involving vesicular structures (24, 25). Future experiments will target specific sphinganine- and sphingosine-derived lipids to correlate loss of HIV-1 infectivity with specific lipids and to determine their mechanism of action. Our results with FB1 suggest that lipid active drugs effectuating only subtle changes in membrane composition have the potential to become effective therapeutic agents against HIV.
Lipids of Native HIV-1 Membranes and DRM.
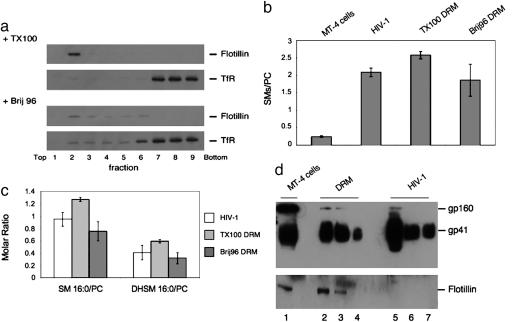
Our analysis of HIV-1 lipid composition revealed a similar picture as previously reported for DRM, and we therefore performed a direct comparison of HIV-1 and DRM from infected MT-4 cells. DRM were prepared by using the detergents Triton X-100 and Brij 96, which differ in their ability to solubilize specific proteins and lipids, with Triton X-100 being more stringent (26). Fractions after isopycnic buoyant density centrifugation were analyzed regarding distribution of the DRM marker flotillin and of transferrin receptor, which is not associated with DRM (Fig. 4a). Flotillin was restricted to fraction 2 in the case of Triton X-100 extraction, whereas transferrin receptor was found at the bottom of the gradient (fractions 7–9), thus defining fraction 2 as DRM fraction. Brij 96 was much less stringent, resulting in a broader distribution of both flotillin and transferrin receptor (Fig. 4a Lower). Fig. 4b shows that the ratio of SMs/PC was strongly increased in DRM from Triton X-100 as well as from Brij 96-treated cells (compared with total cell lipids) and was similar to native HIV-1 lipids. This result was also observed when the ratios of SM 16:0/PC and DHSM 16:0/PC were analyzed (Fig. 4c), indicating that DHSM was enriched to a similar extent in native HIV-1 membranes and DRM as well. Thus, the HIV-1 membrane shows lipid ratios very similar to DRM, despite being prepared without any detergent treatment.
Fig. 4.
DRM from infected cells and native HIV-1 membranes show similar enrichment of SM and DHSM. HIV-1 particles and DRM were isolated as described in Materials and Methods. The SM, DHSM, and PC content in virus membranes and DRM was determined by nano-electrospray ionization tandem mass spectrometry, whereas the distribution of selected proteins was determined by Western blot. (a) DRM obtained by cold extraction of HIV-1 infected MT-4 cells with Triton X-100 (TX100, Upper) or Brij 96 (Lower) were analyzed for raft (flotillin 1) and nonraft (transferrin receptor, TfR) marker distribution. (b) SMs to PC ratios in total cell membranes, HIV-1, and DRM obtained with TX100 or Brij 96. (c) SM (16:0) and DHSM (16:0) to PC ratios in HIV-1 and DRM. (d) Extracts from infected MT-4 cells (lane 1), DRM from infected cells (obtained by cold TX100 extraction; lanes 2–4), and purified HIV-1 (lanes 5–7) were subjected to Western blot analysis detecting flotillin or gp41. The amount of cell extracts loaded was normalized according to cell number (105 cells in lane 1; DRM from 3 × 105, 2 × 105, and 105 cells in lanes 2, 3 and 4, respectively), and the amount of virus loaded was normalized according to total PC and SM content of DRM and HIV-1 preparations with equal amounts of lipids loaded in lanes 4 and 6 (lane 5, 500% of lane 6; lane 7, 50% of lane 6). Error bars represent standard deviation of the mean.
To compare the segregation of lipids and proteins into DRM and viral membranes, respectively, we analyzed flotillin and the HIV-1 transmembrane glycoprotein (gp)41 (and its precursor gp160) in the various fractions (Fig. 4d). In the case of flotillin, ≈50% was recovered in the DRM fraction from Triton X-100 extracted cells (Fig. 4d Lower, compare lanes 1 and 3). Recovery appeared to be lower for HIV-1 gp41 (Fig. 4d Upper, compare lanes 1 and 3), whereas the uncleaved precursor gp160 was virtually absent from DRM. A very different result was observed for extracts from native HIV-1, where gp41 was strongly enriched (Fig. 4d, compare lanes 4 and 6), and flotillin was not detectable. This finding differs from previous studies reporting the incorporation of other DRM-associated proteins into HIV-1 (for review, see ref. 2). The segregation of flotillin from HIV membranes is consistent with the viral budding platform being formed by coalescence of a subset of preexisting lipid microdomains, which may be induced by the viral structural proteins. Because the native HIV-1 membrane resembles a DRM, we analyzed lipid and protein distribution after isopycnic density gradient centrifugation of Triton X-100 extracts from purified HIV-1 particles. As observed for cell extracts, HIV-1 lipids and proteins segregated into buoyant and nonbuoyant fractions. The distribution into detergent-sensitive and detergent-resistant fractions was similar for SMs and gp41, with approximately one-third in the detergent-resistant fraction. This result indicates that there is no obvious preference of the viral fusion machinery for membrane domains of different detergent sensitivity. The SM 16:0/PC and DHSM 16:0/PC ratios were similar for native HIV-1 membranes, DRM from HIV-1, and DRM from infected cells.
The HIV-1 Lipidome: Its Relation to Lipid Rafts.
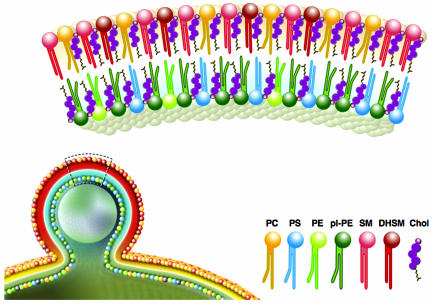
Our quantitative analysis of the lipids of highly purified native HIV-1 allows us to calculate an estimate for the numbers of individual lipid molecules per virus particle, even with respect to their molecular species distribution (the HIV-1 lipidome). We estimate that this analysis includes >95% of all HIV-1 lipids based on prior determination of other membranes (7, 8), and future experiments will be directed at quantifying minor lipid groups (e.g., phosphatidylinositol phosphates). A graphical representation of the lipid composition of HIV-1 membranes compared with the surrounding membrane of the producer cell is shown in Fig. 5. Quantitation of HIV-1 lipids and structural proteins from the same sample provides a quantitative description of lipid molecules per virion. Assuming 4,900 Gag proteins (27), this calculation yielded ≈296,000 lipid molecules per average HIV-1 particle for two independent preparations, with the distribution of lipids shown in Table 2 (for details of calculation, see Supporting Text, which is published as supporting information on the PNAS web site). The number of lipids per virion can also be calculated from the viral diameter and the average footprint of a lipid molecule. With an average outer HIV-1 diameter of 145 nm (28), this calculation yields a theoretical number of 255,000 lipids per particle assuming a 0.5 nm2 surface area per lipid molecule, which is in excellent agreement with the experimentally determined number.
Fig. 5.
The lipid composition of HIV particles. For details, see text.
Table 2.
The lipid composition of HIV-1
| Lipid molecules per average HI virion | |
|---|---|
| PC | 26,000 |
| SM | 37,000 |
| DHSM | 17,000 |
| PE | 13,000 |
| pl-PE | 44,000 |
| PS | 25,000 |
| Chol | 134,000 |
| Cer | 160 |
| HC | 600 |
For details, see Supporting Text.
The observation that the lipid composition of HIV-1 is very similar to that proposed for lipid microdomains (rafts) (29) in at least five different aspects (enriched in saturated lipids, PS, pl-PE, cholesterol, and sphingolipids) strongly supports the hypothesis that HIV-1 buds from cellular membrane microdomains. The absence of the bona fide raft marker flotillin from pure HIV-1 preparations is in line with the concept that HIV budding is a specific raft clustering process, whereas DRM represents general aggregations of raft microdomains. HIV proteins are probably interacting with rafts, and interactions between them may drive the clustering process. It will be a future challenge to identify the viral gene products that determine HIV-1 lipid composition and to define potential host cell differences and their influence on infectivity. Most importantly, however, the lipid composition of highly purified native HIV-1, prepared in the absence of any detergent, is among the strongest evidence for the existence of raft-like lipid microdomains in biological membranes of living cells.
Materials and Methods
Materials.
FB1 and Brij 96 were obtained from Sigma. Sphingosylphosphorylcholine was obtained from Matreya (Pleasant Gap, PA). Unsaturated PC species for transphosphatidylation and SM (porcine brain and chicken egg) were purchased from Avanti Polar Lipids, and fatty acids and Triton X-100 were from Merck.
Antibodies.
Monoclonal mouse anti-human transferrin receptor antibody (clone H68.4) was from Zymed. The polyclonal rabbit anti-flotillin-1 antibody was kindly donated by J. B. Helms (University of Utrecht, The Netherlands). Monoclonal antibody against HIV-1 gp41 was derived from cell culture media of Chessie 8 cells obtained from George Lewis (30) through the National Institutes of Health AIDS research and reagent reference program. Polyclonal rabbit serum against the HIV-1 capsid protein had been raised against purified protein.
Cell Culture and Virus Purification.
MT-4 cells (31) were maintained at 37°C and 5% CO2 in RPMI 1640 medium supplemented with 10% heat-inactivated FCS, antibiotics, 4 mM l-glutamine, and 5 mM Hepes. Cells were infected with HIV-1 strain NL4–3 (32), and the virus was harvested from cocultures of infected and uninfected cells before cytopathic effects were observed as described in ref. 12. For inhibitor treatment, FB1 in DMSO was added at a final concentration of 50 μM.
HIV-1 purification was performed as described (12). Briefly, particles were concentrated from cleared media by centrifugation through a cushion of 20% (wt/wt) sucrose in PBS. Concentrated HIV-1 was further purified by velocity gradient centrifugation on an Optiprep gradient (Axis-Shield, Oslo, Norway). The visible virus band was collected and pelleted yielding a 1,800-fold concentration compared with the initial volume. Virus titers were determined on TZM cells containing a β-galactosidase gene under control of the HIV-1 promoter as described (33).
Quantitative Analysis of HIV-1 Release.
Virus release was quantitated by ELISA of cleared culture supernatants from infected MT-4 cells detecting the HIV-1 capsid protein p24. Quantitative Western blot analysis was performed by using the Odyssey infrared imaging system from Li-Cor (Lincoln, NE).
Isolation of DRM.
Infected or uninfected MT-4 cells (1 × 107) were washed and extracted on ice for 15 min in 0.3 ml of lysis buffer (50 mM Tris·HCl, pH 7.4/150 mM NaCl/5 mM EDTA/1 mM DTT/1% Triton X-100). The lysate was dounced ten times, mixed with 0.6 ml of Optiprep (Axis-Shield) and overlaid with 2.5 ml 28% Optiprep in lysis buffer, followed by 0.6 ml of lysis buffer in a SW60 tube. Tubes were centrifuged at 126,000 × g for 3 h at 4°C. Eight fractions (450 μl each) were collected from the top. Western blots of gradient fractions were probed with antibodies against flotillin-1, transferrin receptor, or HIV-1 gp41. DRM fractions were directly subjected to mass spectrometer analysis. DRM preparation from HIV-1 particles was as described above, except that purified virus from 600 ml of culture supernatant from infected MT-4 cells was used as input material.
Lipid Analysis.
Lipid extractions were performed according to the method of Bligh and Dyer (34). Quantitation of PC, SM, DHSM, and cholesterol was performed as described (35). HC and Cer scanning was performed by precursor ion scanning for fragment ion 264 Da (positive ion mode) at a collision energy of 35 eV (1 eV = 1.602 × 10−19 J) (HC) or 30 eV (Cer). Quantitative data given refer to major species N-palmitoyl-HC and N-palmitoyl-Cer, respectively. Quantitation of PE and PS was performed by neutral loss scanning, selecting for a neutral loss of 141 Da or 185 Da (positive ion mode), respectively, with a collision energy of 20 eV. pl-PE quantitation was performed by precursor ion scanning for fragment ion m/z 196 Da (negative ion mode; collision energy of 40 eV). Unsaturated PE and PS standards were synthesized and purified via HPLC as described (36). Quantitative analyses were performed as described (13, 37). Phosphate determination was performed according to Rouser (38).
Characterization of DHSM.
See Supporting Text.
Supplementary Material
Acknowledgments
We thank Walter Nickel [Heidelberg University Biochemistry Center (BZH)] for helpful comments and critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grants SPP1175 (to B.B. and F.T.W.) and SFB638, A9 (to H.-G.K.).
Abbreviations
- Cer
ceramide
- DHSM
dihydrosphingomyelin
- DRM
detergent-resistant membrane
- FB1
fumonisin B1
- HC
monohexosylceramide
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- SM
sphingomyelin.
Footnotes
Conflict of interest statement: The communicating member K.S. is a founder in a small start-up company called Jado Technologies that has raft technology as its specialty.
References
- 1.Simons K., Vaz W. L. Annu. Rev. Biophys. Biomol. Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 2.Ono A., Freed E. O. Adv. Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- 3.Kusumi A., Koyama-Honda I., Suzuki K. Traffic. 2004;5:213–230. doi: 10.1111/j.1600-0854.2004.0178.x. [DOI] [PubMed] [Google Scholar]
- 4.Simons K., Toomre D. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 5.Munro S. Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 6.Lagerholm B. C., Weinreb G. E., Jacobson K., Thompson N. L. Annu. Rev. Phys. Chem. 2005;56:309–336. doi: 10.1146/annurev.physchem.56.092503.141211. [DOI] [PubMed] [Google Scholar]
- 7.Aloia R. C., Jensen F. C., Curtain C. C., Mobley P. W., Gordon L. M. Proc. Natl. Acad. Sci. USA. 1988;85:900–904. doi: 10.1073/pnas.85.3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aloia R. C., Tian H., Jensen F. C. Proc. Natl. Acad. Sci. USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ono A., Freed E. O. Proc. Natl. Acad. Sci. USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y. E., Kielian M. J. Virol. 2000;74:7708–7719. doi: 10.1128/jvi.74.17.7708-7719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheiffele P., Rietveld A., Wilk T., Simons K. J. Biol. Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 12.Welker R., Hohenberg H., Tessmer U., Huckhagel C., Kräusslich H. G. J. Virol. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brügger B., Erben G., Sandhoff R., Wieland F. T., Lehmann W. D. Proc. Natl. Acad. Sci. USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Faure J., Blanc N. S., Matile S., Dubochet J., Sadoul R., Parton R. G., Vilbois F., Gruenberg J. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 15.Fridriksson E. K., Shipkova P. A., Sheets E. D., Holowka D., Baird B., McLafferty F. W. Biochemistry. 1999;38:8056–8063. doi: 10.1021/bi9828324. [DOI] [PubMed] [Google Scholar]
- 16.Sullards M. C. Methods Enzymol. 2000;312:32–45. doi: 10.1016/s0076-6879(00)12898-8. [DOI] [PubMed] [Google Scholar]
- 17.Byrdwell W. C., Borchman D., Porter R. A., Taylor K. G., Yappert M. C. Invest. Ophthalmol. Vis. Sci. 1994;35:4333–4343. [PubMed] [Google Scholar]
- 18.Epand R. M. Biophys. J. 2003;84:3102–3110. doi: 10.1016/S0006-3495(03)70035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuikka M., Ramstedt B., Ohvo-Rekila H., Tuuf J., Slotte J. P. Biophys. J. 2001;80:2327–2337. doi: 10.1016/S0006-3495(01)76203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nyholm T., Nylund M., Soderholm A., Slotte J. P. Biophys. J. 2003;84:987–997. doi: 10.1016/S0006-3495(03)74915-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagan N., Zoeller R. A. Prog. Lipid Res. 2001;40:199–229. doi: 10.1016/s0163-7827(01)00003-0. [DOI] [PubMed] [Google Scholar]
- 22.Campbell S. M., Crowe S. M., Mak J. AIDS. 2002;16:2253–2261. doi: 10.1097/00002030-200211220-00004. [DOI] [PubMed] [Google Scholar]
- 23.Wang E., Norred W. P., Bacon C. W., Riley R. T., Merrill A. H., Jr. J. Biol. Chem. 1991;266:14486–14490. [PubMed] [Google Scholar]
- 24.Stüven E., Porat A., Shimron F., Fass E., Kaloyanova D., Brügger B., Wieland F. T., Elazar Z., Helms J. B. J. Biol. Chem. 2003;278:53112–53122. doi: 10.1074/jbc.M300402200. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Thiele C., Huttner W. B. Traffic. 2000;1:952–962. doi: 10.1034/j.1600-0854.2000.011205.x. [DOI] [PubMed] [Google Scholar]
- 26.Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K. Proc. Natl. Acad. Sci. USA. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briggs J. A., Simon M. N., Gross I., Kräusslich H. G., Fuller S. D., Vogt V. M., Johnson M. C. Nat. Struct. Mol. Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 28.Briggs J. A., Wilk T., Welker R., Kräusslich H. G., Fuller S. D. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pike L. J., Han X., Chung K. N., Gross R. W. Biochemistry. 2002;41:2075–2088. doi: 10.1021/bi0156557. [DOI] [PubMed] [Google Scholar]
- 30.Abacioglu Y. H., Fouts T. R., Laman J. D., Claassen E., Pincus S. H., Moore J. P., Roby C. A., Kamin-Lewis R., Lewis G. K. AIDS Res. Hum. Retroviruses. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 31.Harada S., Koyanagi Y., Yamamoto N. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 32.Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller B., Daecke J., Fackler O. T., Dittmar M. T., Zentgraf H., Kräusslich H. G. J. Virol. 2004;78:10803–10813. doi: 10.1128/JVI.78.19.10803-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bligh E. G., Dyer W. J. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 35.Brügger B., Graham C., Leibrecht I., Mombelli E., Jen A., Wieland F., Morris R. J. Biol. Chem. 2004;279:7530–7536. doi: 10.1074/jbc.M310207200. [DOI] [PubMed] [Google Scholar]
- 36.Koivusalo M., Haimi P., Heikinheimo L., Kostiainen R., Somerharju P. J. Lipid Res. 2001;42:663–672. [PubMed] [Google Scholar]
- 37.Brügger B., Sandhoff R., Wegehingel S., Gorgas K., Malsam J., Helms J. B., Lehmann W. D., Nickel W., Wieland F. T. J. Cell Biol. 2000;151:507–518. doi: 10.1083/jcb.151.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouser G., Fleischer S., Yamamoto A. Lipids. 1970;5:494–496. doi: 10.1007/BF02531316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.