Abstract
The conformation of DNA molecules tethered to the surface of a microarray may significantly affect the efficiency of hybridization. Although a number of methods have been applied to determine the structure of the DNA layer, they are not very sensitive to variations in the shape of DNA molecules. Here we describe the application of an interferometric technique called spectral self-interference fluorescence microscopy to the precise measurement of the average location of a fluorescent label in a DNA layer relative to the surface and thus determine specific information on the conformation of the surface-bound DNA molecules. Using spectral self-interference fluorescence microscopy, we have estimated the shape of coiled single-stranded DNA, the average tilt of double-stranded DNA of different lengths, and the amount of hybridization. The data provide important proofs of concept for the capabilities of novel optical surface analytical methods of the molecular disposition of DNA on surfaces. The determination of DNA conformations on surfaces and hybridization behavior provide information required to move DNA interfacial applications forward and thus impact emerging clinical and biotechnological fields.
Keywords: hybridization, microarray, spectroscopy
DNA array technology has become a widespread tool in biological research with applications in expression screening, sequencing, and drug discovery, all benefiting greatly by the highly paralleled detection of the technique. One of the defining characteristics of a DNA array is the availability of the single-stranded probes for hybridization with the target. Immobilized molecules located farther away from the solid support are closer to the solution state and are more accessible for contact with dissolved analytes. The surface, especially a hydrophobic one, acts as a shield for probes positioned close to it because of the associated steric factors and lack of diffusion of the bound molecules (1–5). Thus, knowing the physical structure of DNA probes may prove useful not only in the future development and fabrication of microarrays, but also in designing new applications (6).
Recently, advances have been made to characterize the structure of surface-bound DNA probes using such optical or contact methods as ellipsometry, optical reflectivity (7, 8), neutron reflectivity (9), x-ray photoelectron spectroscopy (10), FRET (11, 12), SPR (13, 14), and AFM (15–18). This previous work has helped in visualizing the structure of the surface-bound DNA depending on its density and surface charge. However, most experimental techniques characterize the DNA layer as a single entity (for instance, parameterizing its thickness or density) without examining the specific positions of internal elements of the DNA chain. Spectral self-interference fluorescence microscopy (SSFM) can measure the vertical position of a fluorescent label above an optically structured silicon chip. We show that locating the label attached to a certain position within a DNA chain provides insight into the shape of DNA molecules bound to the surface.
Results
We study the conformation of single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) on glass surfaces by using 50- and 21-nt oligonucleotides. In all studies, the first strand of the DNA was covalently bound to the surface of an oxide-coated silicon substrate at its 5′ end. Experiments were performed with fluorescein markers bound to either the first strand at its distal 3′ end, or the second strand at its 3′ or 5′ end. Below, we describe the detection principle along with results on ssDNA and dsDNA conformation. A summary of all of the data is presented in Table 1.
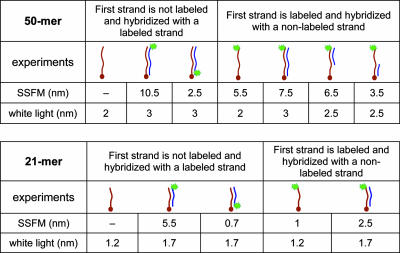
Table 1.
Average height of the fluorescent marker above the surface as measured by fluorescence interference (SSFM) and the optical thickness of the DNA layer on the surface (measured by WL reflectance)
In all experiments we start with a ssDNA covalently bound to the surface (shown on the left in red). In one set of experiments, the first oligonucleotide (either 50- or 21-mer) is covalently bound to the surface and does not have a fluorescent label. In this case, only WL measurements can be performed prior to hybridization. After hybridization with a fluorescently labeled matching strand (shown on the right in blue), SSFM yields the average fluorophore height attached to either proximal or distal end of the resulting dsDNA. In the second set of experiments, covalently bound oligonucleotide (either 50-or 21-mer) is fluorescently labeled at the distal end and SSFM yields the average height of the ssDNA. When a second, unlabeled matching strand is hybridized, the DNA layer now consists of two species: the dsDNA and the unhybridized strands both of which are carrying a fluorescent marker, but expected at different heights. SSFM measures the average height of the fluorophores. Experiments with 50-mers also include when the first strand is annealed with a 21-mer complementary to either its top or bottom part as schematically shown. The values shown are averages from multiple measurements. Whereas the accuracy of a single height measurement is ≈0.2 nm, the variation between different measurements is ≈10% of the nominal values.
Detection Principle.
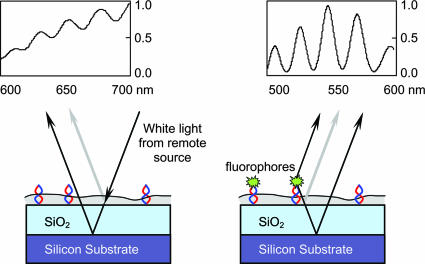
We use a complementary combination of a traditional reflection technique and an interferometric fluorescence spectroscopy technique to determine the average optical thickness of biological layers as well as the height of sparse fluorescent markers, both with subnanometer accuracy. Samples are prepared on layered dielectric films, specifically an oxide-coated silicon substrate. The first technique, white light (WL) reflection spectroscopy, is based on spectral variations of reflection from thin transparent films. Interference of light reflected from the top surface and a buried reference surface results in periodic oscillations in the reflection spectrum as shown in Fig. 1. The principle is similar to the interference-based detection technique using color variations due to increased path length as a consequence of surface binding on optically coated silicon (19). Our spectral measurements obtain very high accuracy (≈0.2 nm), comparable to ellipsometry. The result is an average optical density measurement of the biological film and thus provides a precise relative measure of the additional “optical” mass on the substrate, after, e.g., hybridization.
Fig. 1.
Schematic representation of the interferometric experimental configurations are shown along with examples of measured spectra for each technique. (Left) WL reflection spectroscopy is based on spectral variations of reflection from thin transparent films. Interference of light reflected from the top surface and a buried reference surface results in periodic oscillations. (Right) The SSFM technique maps the spectral oscillations emitted by a fluorophore located on a layered reflecting surface into a precise position determination. Analysis of the spectral oscillations (using a grating spectrometer) due to the self-interference from the direct and reflected emission yields the vertical position of that fluorophore with subnanometer accuracy.
The second technique, SSFM (20), is an interferometric technique in fluorescent imaging that analyzes the spectral oscillations emitted by a fluorophore located on a layered reflecting surface and yields a precise position determination (Fig. 1). The spectral oscillations are due to the self-interference from the direct and reflected emission and thus encode the vertical position of that fluorophore with subnanometer accuracy. These markers can be sparse or buried under a biological film. In contrast to earlier fluorescence interference microscopy techniques that rely on intensity variation of total fluorescent emission (21, 22), SSFM utilizes spectral information and provides higher precision with a single measurement. The following discussion of measurements on oligonucleotides illustrates the complementary features and capabilities of these spectroscopies.
WL Reflectivity Measurements.
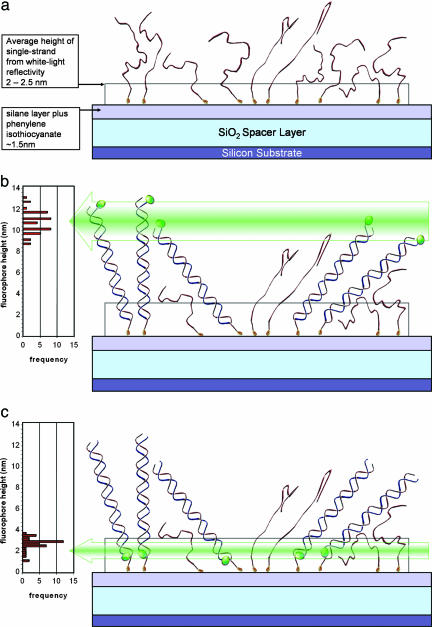
As schematically shown in Fig. 2, oligonucleotides carrying a 5′-amino tag are covalently bound to an aminated surface via a homobifunctional crosslinker. Using WL reflectivity, we determine progressive growth of the surface-bound thin films during DNA immobilization steps. The thickness of the silane layer is 0.8–1.0 nm, which roughly corresponds to a monolayer; phenylene isothiocyanate adds another 0.5–0.6 nm. Immobilization of DNA leads to a further increase in the optical thickness. In this particular experiment, oligonucleotides are not fluorescently labeled, and the average film thickness determined by WL reflection spectroscopy is depicted as transparent box above the oxide in Fig. 2a.
Fig. 2.
WL and fluorescence interference measurements schematically shown for ssDNA and dsDNA immobilized on a flat surface. Nonlabeled ssDNA covalently bound on the surface (a, top) after hybridization with complementary strand labeled at the 5′ (distal) end (b) and 3′ (proximal) end (c, bottom).
The physical thickness of the DNA layer can be established from WL reflectivity or ellipsometry only if the value of the index of refraction is known. In these experiments, we assumed the index of refraction of DNA to be 1.46 as was measured for dense layers (23). We measured the optical film thickness for ssDNA of 21 and 50 nt as 1.0–1.5 and 2.0–2.5 nm, respectively. Although the precision of absolute thickness will depend on the index, WL reflectivity provides an accurate relative measure of additional mass on the surface and can thus monitor the efficiency of hybridization. For example, adding complementary second strands to 50-mers results in an increase in the film thickness by ≈1.0 nm (compared with ≈2 nm for the first strand) corresponding to a hybridization efficiency of ≈50%. Precise determination of the hybridization efficiency is not crucial for the significant conclusions drawn about the conformation of ssDNA and dsDNA on surfaces. An approximate value is estimated to check for consistency of independent measurements. Incomplete hybridization is schematically illustrated by converting half of the oligonucleotides from Fig. 2a to dsDNA in Fig. 2 b and c.
Determination of the Position of a Fluorescent Label.
Once the optical thicknesses of the layers are established, they can be used in SSFM measurements to determine the position of the fluorophores relative to the surface.
The first experiment studied the elevation of 21- and 50-bp dsDNA fragments. In principle, the maximum elevation of the label is limited by the length of the double helix, which is ≈7 nm for 21-bp and ≈17 nm for 50-bp fragments. dsDNA has a persistence length of ≈50 nm (24), so the short fragments in our experiments can be viewed as rigid rods on hinges. A simple rod, hinged to the surface, would have an average height of the distal label, assuming free rotation of one half of the length, with an average tilt angle of 60° from normal. However, free rotation may be limited by steric constraints from nearby DNA molecules and interactions with the surface. Using SSFM, we measured the elevation of the label on top of DNA double helices to be 5.5 nm for 21-bp fragments and 10.5 nm for 50-bp fragments, which represents tilt angles from the normal of 50° and 40°, respectively. These values are a measure of the average distribution of heights within the microscope focal spot. Whereas some DNA helices may be standing straight up, others may even be lying flat on the surface. In Fig. 2b, we also show statistical distribution of many measurement results illustrating a variance of the average label position on the order of 1 nm.
If the label on the second strand is at the 3′ end, its location in the double helix will be at the bottom, close to the surface. Although we did not expect to see a significant variation, the proximal, 3′-end label on 50-bp DNA is elevated by ≈2–3 nm compared with only 0.5–1 nm in the case of the 21-bp fragment. Theoretically, the position of the proximal label on stand-alone double-stranded fragments, especially sharing the sequence of the first 21 bp, should not be higher just because the double-stranded fragment above it is longer. However, this may be due to the fact that a 50-mer is long enough to have stable partial hybridization that could free the proximal end and yield an elevated label position. Steric hindrance is also a possibility.
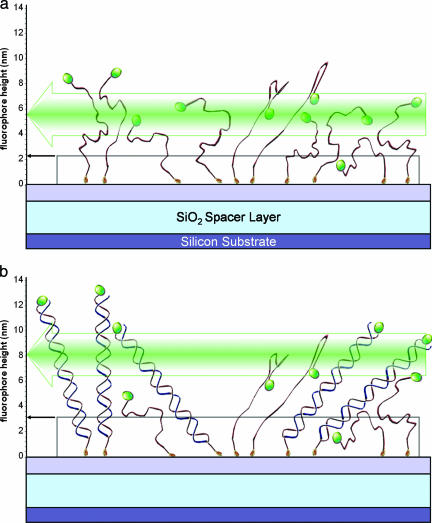
We also studied the conformation of ssDNA by measuring the height of fluorescent tags attached to the free end of surface-bound DNA oligonucleotides (Fig. 3.). Unlike dsDNA, ssDNA is flexible and little is known about the shape or size of ssDNAs on the surface. AFM measurements suggest that ssDNA immobilized on a surface exists in a globular conformation (16). However, there are reports that because of steric hindrance from nearby molecules, ssDNA may change its conformation from a random coil to more extended forms (23, 25). The fluorescent label attached to the distal end of surface-bound single-stranded 21-mer is found to be close to the surface: within 1 nm. In a similar situation, 50-mers show much higher location of the label: 5.5 nm above the surface as illustrated in Fig. 3b. It is difficult to calculate the average expected position of an end-label for random coils of this size, yet the disproportionally high location of the end-label on 50-mers points out to a considerably more extended conformation compared with 21-mers. This extended conformation may be caused by the steric effect from closely located grafts in the DNA layer. The surface density of immobilized ssDNA measured by using a radiolabel is ≈35 fmol/mm2 for both 21- and 50-mers, which translates into 11-nm distances between adjacent molecules or a 5.5-nm radius of free space around each. At the same time, the length of a fully extended 50-mer is 27.5 nm, enough to interact with its neighbors, at least intermittently. It appears that there is a pronounced effect from nearby molecules that we observe only with 50-mers and not with 21-mers.
Fig. 3.
WL and fluorescence interference measurements schematically shown for ssDNA and dsDNA immobilized on a flat surface. ssDNA labeled at the 3′ (distal) end and covalently bound on the surface (a, top), and after hybridization with complementary (nonlabeled) strand (b).
When a second, unlabeled strand is hybridized, the label at the distal end of the newly formed duplex extends out as well. Unlike the dsDNA in Fig. 2b, the DNA layer now consists of two species: the dsDNA and the unhybridized strands, both of which are carrying a fluorescent marker but at different heights. The average position of the marker should be somewhere in between depending on the degree of hybridization. The binding efficiency can be calculated from comparing the average height of the fluorescent layer above the surface for the two cases where either the first strand or the second strand is labeled. In our experiments, an estimate of the extent of hybridization is between 30% and 50%. This rough estimate is close to the result obtained by WL reflectivity: 50% hybridization for both 21- and 50-mers, demonstrating self-consistency. As a further check, we have performed density measurements with radiolabeled DNA and found it consistent with 50% hybridization. Because of intrinsic limitations such as substrate related quenching of radiative emission, we do not consider the radiolabeling for absolute determination of DNA densities, but rather only for relative estimation of DNA densities.
Interesting results were obtained when we assessed how the conformation of a surface-bound 50-nt labeled oligonucleotide changes when it is annealed with a 21-mer complementary to either its top or bottom part. When a 21-mer complementary to the top section was annealed, the position of the distal end increased from 5.5 to 6.5 nm. However, a different situation is observed when an ssDNA–dsDNA construct has the double-stranded part proximal to the surface. In this case, the position of the label is lower than that of an unhybridized oligo, decreasing in average height from 5.5 to 3.5 nm. Recalling that measurements such as these of pretagged oligos always average over the unhybridized single strands, our data suggests that the distal bound 21-mer construct is nearly vertical, and the proximal bound construct has formed a rotation point allowing the flexible distal end to approach closer to the surface. The sequence of our oligonucleotides rules out the possibility of intramolecular DNA structures.
Table 1 summarizes the entire range of experiments, both with single oligonucleotides and complementary pairs of 21- and 50-bp units, and the final results of hybridization with segments of different length.
Discussion
We have demonstrated that SSFM is a powerful tool for studying the conformation of DNA molecules immobilized on a surface. The method is noncontact, and there are no limitations to applying it to DNA arrays submerged in an aqueous buffer. The technique specifically determines the axial position of the labeled nucleotide only and is therefore complementary to WL reflectivity and ellipsometry techniques. Because the fluorophore height is encoded only through the phase and frequency of the oscillations from the spectral interference, the measurement is insensitive to the intensity of fluorophore emission. Thus, photobleaching, surface density-dependent dye–dye nonradiate transfer effects, or spectral modifications will not affect the results. They may result in an overall intensity variation but will yield similar oscillatory behavior of the spectra; hence, misinterpretation of the height measurement is unlikely. These processes merely alter the total number of photons, and integration times can be easily modified to provide whatever signal-to-noise is required for the necessary precision of the measurement. Details of the methods used for processing spectral data are described below.
A broad range of industries utilizing array technologies involve grafted DNA segments on surfaces with partially hybridized molecules, and yet the majority of experimental techniques are not able to determine ssDNA and dsDNA conformation. The SSFM technique will allow the study of a variety of surface-bound DNA structures, for example, hairpin loops formed with DNA synthesis techniques. The use of hairpin loops offers the potential of a cost-effective method for producing high-quality synthesized dsDNA arrays for protein arrays and transcription factor measurements. The idea is to synthesize arrays of long single strands on the surface whose end sequences include their own complement such that the completed strand is likely to hybridize itself, thereby eliminating the need for batch-synthesized oligonucleotides and on-chip polymerization. Then, by simply tagging the distal end, SSFM can be used to confirm the array completion because the fluorescent label for the fully self hybridized DNA will go down to the surface.
Little is known about the conformation of ssDNAs, and there is no reliable model for predicting the shape and diameter of the random coil form (12). In the absence of base-pairing, long ssDNA has been considered as a flexible polymeric molecule and modeled as a freely jointed chain and a worm-like chain (26). Using SSFM, we estimated the shape of single-stranded pieces of DNA 21 and 50 nt long. In our experiments, energy transfer and quenching have no effect due to intensity insensitivity. Therefore, SSFM can be extended to multiple tags in different spectral windows.
Materials and Methods
Materials.
All oligonucleotides were custom-synthesized by Integrated DNA Technologies (Coralville, IA). The following chemicals were used: acetone (HPLC grade; Merck); 3-aminopropyltriethoxysilane (98%; Aldrich), 1,4-phenylene diisothiocyanate (>98%; Fluka), and dimethyl sulfoxide (anhydrous 99.9%; Aldrich). TE buffer was composed of 10 mM Tris·HCl and 1 mM EDTA. The surfaces were silicon wafers with an ≈5-μm-thick layer of silicon oxide grown by plasma-enhanced chemical vapor deposition and polished chemo-mechanically to a roughness of ≈2 nm. The relatively thick layer of silicon oxide serves as a transparent spacer separating the emitters from the principal reflecting surface, silicon. At larger distances, even a small shift in wavelength leads to sharp changes in the intensity of emitted light. The result was a shorter distance between interference peaks, about four to five peaks in the typical emission spectrum of an organic dye located 10–15 wavelengths above the mirror (20).
Covalent Attachment of DNA to the Surface.
Wafers were cut into 15 × 5-mm chips. The chips were washed with acetone, sonicated in water for 10 min, and cleaned with 10% NaOH for 10 min. The treatment with alkali etched ≈1 nm of silicon oxide without affecting the roughness of the surface. After extensive washing with deionized water, the chips were blown dry, treated with 5% aminopropyltriethoxysilane in acetone for 2 min, washed a few times with acetone, and dried in a vacuum oven at 110°C for 20 min. Longer silanation times cause polymerization of silane and deposition of multiple silane layers, which creates a nonuniform surface and is not suitable for our experiments (27). Aminated chips were functionalized with a homobifunctional crosslinker (1 mg/ml phenylene diisothiocyanate in DMSO, 1-h reaction under argon while stirring), washed a few times with DMSO, and rinsed with water. The chips were then immediately covered with a 10 μM solution of amino-labeled oligonucleotide in 1 M potassium phosphate (pH 8.0) and left for 1 h on a shaker. After washing three times with TE buffer (pH 7.0) containing 1 M NaCl, the chips were either used for measurements or hybridized with a complementary oligo (10 μM in 1 M NaCl/TE, pH 7.0). After hybridization, the chips were washed again three times in the same buffer. The 21-mer oligonucleotide sequence was 5′-GAA TTC GAG CTC GGT ACC CGG-3′; the 50-mer oligonucleotide sequence was 5′-GAA TTC GAG CTC GGT ACC CGG GGA TCC TCT AGA GTC GAC CTG CAG GCA TG-3′. We designed and checked the oligos to have no self-complementarity. The sequences were taken from a polylinker region of a popular cloning vector. For experiments with ssDNA, the oligos were carrying an amino group with a C6 spacer on the 5′ end and an Oregon green 488 label on the 3′ end. For experiments with dsDNA, these had only the amino group. The complementary strand was labeled with Oregon green 488 at either the 5′ or the 3′ end.
Acquisition and Processing of Spectra.
The spectra were obtained with a Renishaw 1000B micro-Raman spectrometer coupled with a Leica DM/LM upright microscope. A low-numerical-aperture objective (×5, 0.12 numerical aperture) was used to minimize the collection cone. An 1,800-grooves-per-mm grating was used with a spectral resolution of 2 cm−1 at 500 nm. For WL measurements, normal Koehler illumination with a standard halogen lamp was used. The light source for fluorescent measurements was the 488-nm line of an argon ion laser. Both WL reflectivity and fluorescence self-interference spectra were fitted by using a custom-built MATLAB application that separates the oscillatory component from the envelope function. This program automatically calculates the parameters of the system such as the thickness of thin films or position of the emitters above the mirror. There is a noticeable variation in the index of refraction of silicon oxide within the wavelength span we used. This variation was taken into account in the fitting algorithm.
The classical model of fluorescence self-interference is described in detail in ref. 28. The essence of the method is that fluorescent emission near reflecting surfaces is modified by the interference between the direct and the reflected waves. The position of the emitter above the mirror has a direct effect on the phase of the resulting oscillatory component and can be deduced from the emission spectra. The model also takes into account the complex reflectivity of the underlying stack of dielectrics and the orientation of the dipole moments of the emitters. Because the curve-fitting algorithm extracts the oscillatory term to determine the label height, our measurements are immune not only to potential quenching of the entire spectrum, but also to any spectral modifications or nonradiative transfer effects, because such effects would result in a similar oscillatory behavior of the spectra.
WL reflectivity measures the optical thickness of the transparent material on top of a mirror. It is a process similar to ellipsometry, except that it works by fitting the spectra of WL reflected from the surface and modified by thin-film interference (20). Measurements were taken from 10 to 20 spots on a chip, usually in a linear pattern. The spot size was determined to be on the order of 10–20 μm.
Quantitation of the Density of Immobilized DNA.
DNA oligonucleotides were radiolabeled at the 3′ end with [α-32P]ddATP (Amersham Pharmacia) by using terminal transferase (Roche). Labeled oligonucleotides were purified on a gel-filtration column packed with Sephadex G-50 gel-filtration resin. DNA immobilization was carried out the same way as for fluorescently labeled oligos. The chips were dried, and the amount of radioactive material on the surface was measured in a scintillation counter.
Nonspecific binding was checked by using radiolabeled oligonucleotides lacking an amino tag and it was found to be negligibly small, ≪1 fmol/mm2.
Acknowledgments
We thank A. Yönet for help with graphical visualization. This work was supported by National Science Foundation Grant DBI0138425, Air Force Office of Scientific Research Grant MURI F-49620-03-1-0379, and National Institutes of Health–National Institute of Biomedical Imaging and BioEngineering Grant 5R01 EB00 756-03.
Abbreviations
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
- SSFM
spectral self-interference fluorescence microscopy
- WL
white light.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Shchepinov M. S., Case-Green S. C., Southern E. M. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Southern E., Mir K., Shchepinov M. Nat. Genet. 1999;21:5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- 3.Mir K. U., Southern E. M. Nat. Biotechnol. 1999;17:788–792. doi: 10.1038/11732. [DOI] [PubMed] [Google Scholar]
- 4.Vainrub A., Pettitt B. M. Phys. Rev. E. 2002;66:041905. doi: 10.1103/PhysRevE.66.041905. [DOI] [PubMed] [Google Scholar]
- 5.Vainrub A., Pettitt B. M. Biopolymers. 2003;68:265–270. doi: 10.1002/bip.10271. [DOI] [PubMed] [Google Scholar]
- 6.Wong K.-Y., Pettitt B. M. Theor. Chem. Acc. 2001;106:233–235. [Google Scholar]
- 7.Chrisey L. A., Lee G. U., O’Ferrall C. E. Nucleic Acids Res. 1996;24:3031–3039. doi: 10.1093/nar/24.15.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray D. E., Case-Green S. C., Fell T. S., Dobson P. J., Southern E. M. Langmuir. 1997;13:2833–2842. [Google Scholar]
- 9.Levicky R., Herne T. M., Tarlov M. J., Satija S. K. J. Am. Chem. Soc. 1998;120:9787–9792. [Google Scholar]
- 10.Herne T. M., Tarlov M. J. J. Am. Chem. Soc. 1997;119:8916–8920. [Google Scholar]
- 11.Charreyre M.-T., Tcherkasskaya O., Winnik M. A. Langmuir. 1997;13:3103–3110. [Google Scholar]
- 12.Murphy M. C., Rasnik I., Cheng W., Lohman T. M., Ha T. Biophys. J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterlinz K. A., Georgiadis R. M., Herne T. M., Tarlov M. J. J. Am. Chem. Soc. 1997;119:3401–3402. [Google Scholar]
- 14.Wolf L. K., Gao Y., Georgiadis R. M. Langmuir. 2004;20:3357–3361. doi: 10.1021/la036125+. [DOI] [PubMed] [Google Scholar]
- 15.Kelley S. O., Barton J. K., Jackson N. M., McPherson L. D., Potter A. B., Spain E. M., Allen M. J., Hill M. G. Langmuir. 1998;14:6781–6784. [Google Scholar]
- 16.Shlyakhtenko L. S., Gall A. A., Weimer J. J., Hawn D. D., Lyubchenko Y. L. Biophys. J. 1999;77:568–576. doi: 10.1016/S0006-3495(99)76913-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenigk R., Carles M., Ip N. Y., Sucher N. J. Langmuir. 2001;17:2497–2501. [Google Scholar]
- 18.Möller R., Csáki A., Köhler J. M., Fritzsche W. Nucleic Acids Res. 2000;28:e91. doi: 10.1093/nar/28.20.e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenison R., Yang S., Haeberli A., Polisky B. Nat. Biotechnol. 2001;19:62–65. doi: 10.1038/83530. [DOI] [PubMed] [Google Scholar]
- 20.Swan A. K., Moiseev L., Cantor C. R., Davis B., Ippolito S. B., Karl W. C., Goldberg B. B., Ünlü M. S. IEEE J. Select. Top. Quantum Electron. 2003;9:294–300. [Google Scholar]
- 21.Braun D., Fromherz P. Phys. Rev. Lett. 1998;81:5241–5244. [Google Scholar]
- 22.Parthasarathy R., Groves J. T. Cell Biochem. Biophys. 2004;41:391–414. doi: 10.1385/CBB:41:3:391. [DOI] [PubMed] [Google Scholar]
- 23.Du Q., Vologodskaia M., Kuhn H., Frank-Kamenetskii M., Volodogskii A. Biophys. J. 2005;88:4137–4145. doi: 10.1529/biophysj.104.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elhadj S., Singh G., Saraf R. F. Langmuir. 2004;20:5539–5543. doi: 10.1021/la049653+. [DOI] [PubMed] [Google Scholar]
- 25.Steel A. B., Levicky R. L., Herne T. M., Tarlov M. J. Biophys. J. 2000;79:975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Zhou H., Ou-Yang Z. C. Biophys. J. 2001;81:1133–1143. doi: 10.1016/S0006-3495(01)75770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida W., Castro R. P., Jou J.-D., Cohen Y. Langmuir. 2001;17:5882–5888. [Google Scholar]
- 28.Moiseev L., Cantor C. R., Aksun I., Dogan M., Goldberg B. B., Swan A. K., Ünlü M. S. J. Appl. Phys. 2004;96:5311–5315. [Google Scholar]