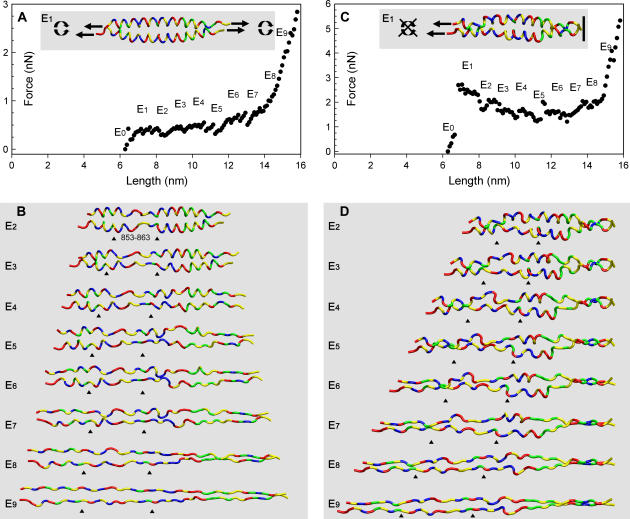
FIGURE 7.
Molecular mechanics simulations of the scallop S2 atomic model allowing rotation (A,B) and disallowing rotation (C,D) of chains relative to one another while stretching both chains. (A,C) The simulated force spectra of incremental extension of the S2 by constrained energy minimizations. Note their similarity to the unique shape of the experimental force spectra from the myosin coiled-coil. E1 configuration showing the stretching mode (inset). (B,D) Images of the S2 atomic model during 1-nm incremental stretching. The coiled-coil unravels at nearly random locations along its length. The ends of the coils unfold first, and then the weakly hydrogen bonded region between residues 853–863 unfolds (E2), followed by the rest of the coils. The position of the 853–863 region at the different extension lengths is marked by black arrowheads. With rotation allowed (A, B), the coiled-coils twists during the extension. With rotation disallowed (C, D), kinks form in the coils. The chains are shown as tubes with color-coded residues: red, acidic; blue, basic; yellow, hydrophobic; green, polar. The E0 configuration is the same as that in Fig. 3. E1–E9 represent structures from 1- to 9-nm extension.