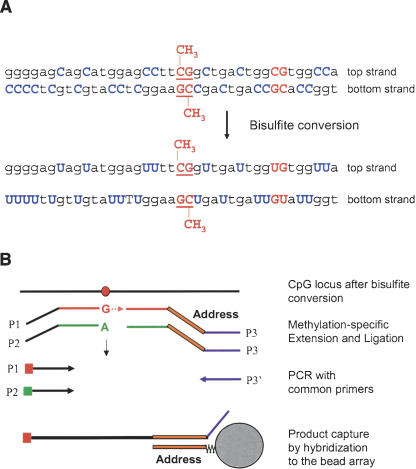
Figure 1.
DNA methylation assay scheme. (A) Bisulfite conversion of DNA. (B) For each CpG site, two pairs of probes were designed to interrogate either the top or bottom strand: an allele-specific oligonucleotide (ASO) and locus-specific oligonucleotide (LSO) probe pair for the methylated state of the CpG site and a corresponding ASO-LSO pair for the unmethylated state. Each ASO consists of a 3′-portion that hybridizes to the bisulfite-converted genomic DNA, with the 3′-base complementary to either the “C” or “T” allele of the targeted CpG site, and a 5′-portion that incorporates a universal PCR primer sequence P1 or P2. The LSOs consist of three parts: At the 5′-end is a CpG locus-specific sequence; in the middle is an address sequence, complementary to a corresponding capture sequence on the array; and at the 3′-end is a universal PCR priming site (P3). Pooled assay oligonucleotides were first annealed to bisulfite-converted genomic DNA. An allele-specific primer extension step was then carried out; ASOs were extended only if their 3′-base was complementary to their cognate CpG site in the gDNA template. Allele-specific extension was followed by ligation of the extended ASOs to their corresponding LSOs, to create PCR templates. The ligated products were then amplified by PCR using common primers P1, P2, and P3′, and hybridized to a microarray bearing the complementary address sequences. P1 and P2 were fluorescently labeled, each with a different dye, and associated with the “T” (unmethylated) allele or the “C” (methylated) allele, respectively.