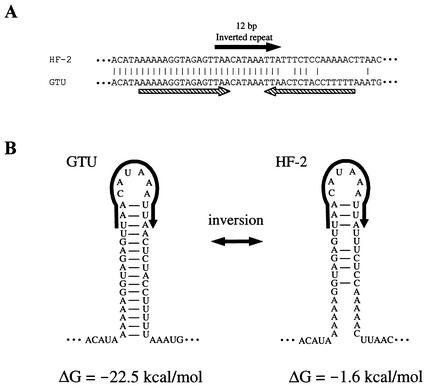
FIG. 5.
Structure of 16-bp inverted-repeat sequence located upstream of p42 gene. (A) Nucleotide sequences of 16-bp inverted-repeat regions from strains HF-2 and GTU. The nucleotide sequence corresponds to nucleotides 63 to 110 in Fig. 4A. The solid arrow indicates a 12-bp inverted-repeat sequence adjacent to a 135-bp inverted DNA sequence. The hatched arrows indicate the 16-bp inverted repeat that is formed only in the GTU sequence. (B) Hairpin structures of the 16-bp inverted repeat. Formation of the hairpin structure is disrupted in the HF-2 sequence by DNA inversion between 12-bp inverted-repeat sequences. The hairpin structures are shown as RNA sequences. The calculated ΔGs of these hairpin structures are shown at the bottom.