Abstract
The hypothesis that host cell surface heparan sulfate is required to promote chlamydial infection was tested using a cell line (CHO-18.4) containing a single retroviral insertion and the concomitant loss of heparan sulfate biosynthesis. Tests of chlamydial infectivity of heparan sulfate-deficient CHO-18.4 cells and parental cells, CHO-22, demonstrated that both were equally sensitive to infection by Chlamydia trachomatis serovars L2 and D. These data do not support the hypothesis and demonstrate that host cell surface heparan sulfate does not serve an essential functional role in chlamydial infectivity.
Investigations of chlamydial infection of mammalian host cells have produced several published studies that have utilized Chinese hamster ovary (CHO) cell mutants deficient in the synthesis of heparan sulfate and/or chondroitin sulfate. CHO-677 is deficient in the synthesis of heparan sulfate because it lacks both N-acetylglucosaminyltransferase and glucuronosyltransferase activities necessary for polymerization of heparan sulfate chains but produces increased amounts of chondroitin sulfate (16). CHO-745 is deficient in the synthesis of both heparan sulfate and chondroitin sulfate because it lacks xylosyltransferase activity required for the initiating step in glycosaminoglycan synthesis (6). CHO-761 lacks galactosyltransferase I for the second step in glycosaminoglycan synthesis and like CHO-745 does not produce heparan sulfate and chondroitin sulfate (7).
In the report by Zhang and Stephens (26) that investigated the functional role of heparan sulfate and chlamydial infectivity in vitro, it was proposed that heparan sulfate-like molecules bound to chlamydial surfaces and bridged binding to host cells, resulting in infection independent of host cell surface heparan sulfate. Consistent with this hypothesis, only small reductions (ca. twofold) in susceptibility to chlamydial infection were observed for heparan sulfate-deficient CHO-761 cells (26). Larger reductions were observed for CHO-677 and CHO-745 cells (four- to eightfold); nevertheless, like the parental cell line CHO-K1, each cell line can readily be >90% infected using static inoculation (18). It was concluded that the reductions in infectivity for these CHO cell mutants were less than would be expected with the loss of a host cell surface molecule essential for the infection process. This reduction in infectivity is the same order of difference between cell lines that synthesize glycosaminoglycans, such as CHO-K1, and HeLa cells that differ in susceptibility by 8- to 10-fold. Other investigators have confirmed the reduction in susceptibility of chlamydiae to these CHO cell mutants but reached different conclusions. Yabushita et al. (24) tested the infectivity of CHO-677 cells using Chlamydia trachomatis serovar L2 and reported a three- to fourfold reduction in infectivity compared to that of CHO-K1 cells. Taraktchoglou et al. (21) found that the infectivity for C. trachomatis serovar L2 was reduced 4.5-fold for CHO-677 cells and 9-fold for CHO-745 cells. Wuppermann et al. (23) tested the infectivity of Chlamydia pneumoniae and reported an eightfold reduction for CHO-677 cells and a fourfold reduction for CHO-745 cells. Su et al. (20) tested the infectivity of the C. trachomatis murine biovar (MoPn) and found a threefold reduction for CHO-677 cells and a fourfold reduction for CHO-745 cells. Each of these authors interpreted the differences in chlamydial infectivity to be sufficient to support the conclusion that host cell surface heparan sulfate-containing proteoglycans are essential for the infectivity of mammalian cells by chlamydiae. Such conclusions are tempting as they appear consistent with the observation that chlamydial infectivity in vitro is very susceptible to inhibition by exogenously added heparin or heparan sulfate (3, 15, 26).
One concern about the use of these CHO cell mutants is that they were selected following chemical mutagenesis that likely introduces multiple mutations in each clone (6, 7, 16). Unfortunately, in none of the chlamydial studies supporting the role of host cell heparan sulfate in chlamydial infectivity had the putative mutation been complemented to demonstrate a recovery of phenotype. Thus, it is not known whether the loss of the cellular ability to synthesize heparan sulfate is implicated as the molecular basis for the relative resistance to infectivity or whether this phenotype is confounded by additional mutations or unexpected outcomes of cellular functions indirectly related to the uncharacterized mutation. Jan et al. (10) derived a CHO cell mutant (CHO-18.4) that is deficient in the production of heparan sulfate and that also produces reduced amounts of chondroitin sulfate. This cell line was derived using a retroviral vector (22) and was shown to have a single retroviral insertion in the genome (10). We hypothesized that if host cell surface heparan sulfate plays an important role in chlamydial infection, then this cell line must be resistant to infection by chlamydiae.
MATERIALS AND METHODS
Organisms and cell lines.
C. trachomatis L2/434/Bu was grown for 48 h in suspension cultures of L929 cells containing approximately 1 × 106 cells/ml. C. trachomatis strain D/UW-3/Cx was grown for 72 h on confluent HeLa 229 cell monolayers (∼5 × 107 cells) in T-150 tissue culture flasks (Costar, Corning, NY). L929, HeLa 229, CHO-K1, CHO(pgsG)-224, CHO-224.GOO, CHO-22, and CHO-18.4 cells were routinely cultured in RPMI 1640, DME H-21, or Ham's tissue culture medium (Gibco, Gaithersburg, MD) supplemented with 10% bovine serum (HyClone, Logan, UT), 0.1 mg/ml streptomycin sulfate (Sigma, St. Louis, MO), 0.1 mg/ml vancomycin hydrochloride (Sigma), and 2 mM glutamine (Sigma). CHO-22 and CHO-18.4 cell lines were generously provided by D. E. Griffin, Johns Hopkins University (10). CHO-K1, CHO-224, and CHO-224.GOO cells were the gift of J. Esko, University of California, San Diego (1). For chlamydial growth, the medium was additionally supplemented with 1 μg/ml cycloheximide (Sigma) following infection with chlamydial strains. Elementary bodies (EB) were isolated from infected cell lines by sonication and purified by centrifugation through discontinuous Renografin density gradients (E. R. Squibb & Sons, Princeton, NJ) as previously described (14). Purified EB were washed with sterile Hanks' balanced salt solution (HBSS; Gibco), suspended in sterile sucrose-phosphate-glutamic acid buffer, and frozen at −70°C until needed. Chlamydial EB were enumerated as numbers of inclusion-forming units (IFU) as previously described (4) or by the chlamydial infection. Alternatively, infectivity was quantitated by real-time PCR, wherein infected monolayers were lysed at 72°C for 10 min in buffer containing guanidine-HCl and proteinase K, and total chlamydial and host cell genomic DNA was isolated per the manufacturer's instructions (High Pure; Roche, Indianapolis, IN). Using Chlamydia-specific 16S primers (9), we analyzed the samples by quantitative PCR (Cepheid, Sunnyvale, CA), with 35 cycles of 95°C for 15 s, 55°C for 5 s, and 72°C for 10 s using SYBR green (Roche), 5 mM MgCl2, and 0.5 μM of each primer.
EB binding assay.
CHO-22 or CHO-18.4 cells (∼2 × 105 cells/ml) were plated onto 12-mm glass coverslips placed in 24-well tissue culture plates. After 24 h, CHO cell monolayers were washed twice with phosphate-buffered saline (PBS) (0.1 g/liter CaCl2, 0.1 g/liter MgCl2, 0.2 g/liter KCl, 0.2 g/liter KH2PO4, 8.0 g/liter NaCl, 2.16 g/liter Na2HPO4), and either C. trachomatis serovar L2 or D EB were added in excess to the monolayers and incubated for 30 min at room temperature. Cell monolayers were washed twice with PBS and fixed with 100% methanol for 10 min at room temperature, rinsed with PBS, and stained with fluorescein isothiocyanate (FITC)-conjugated Chlamydia-specific monoclonal antibody. Samples were observed for qualitative differences in the EB associated with cells using a fluorescence microscope and photographed.
EB infectivity assay.
CHO cells (∼2 × 105 cells/ml) were plated in 24-well tissue culture plates. After 24 h, each well was washed two times with PBS, and cells were infected with either C. trachomatis serovar L2 or serovar D EB serially diluted in HBSS. In some experiments, EB were prepared as described above in HBSS containing various concentrations of heparin (0 mg/ml, 0.25 mg/ml, 0.5 mg/ml, or 1.0 mg/ml). The plates were incubated on a rocking platform for 2 h at room temperature. After infection, monolayers were washed twice with PBS and incubated in DME H-21 medium supplemented with 10% bovine serum, 2 mM glutamine, 0.1 mg/ml streptomycin, 0.1 mg/ml vancomycin, and 1.0 μg/ml cycloheximide for 48 h. Infected monolayers were washed, fixed, and stained with FITC-conjugated monoclonal antibody as described above. Inclusions were counted in 10 (20× objective) fields for each dilution in duplicate, and the mean number of inclusions per well was determined. A low multiplicity of infection was tested and analyzed to provide the most accurate estimation of infectivity.
Cellular heparan sulfate detection.
CHO cell monolayers were prepared (CHO-22, CHO-18.4, or a mixture of 90% CHO-18.4 and 10% CHO-22 cells) on 12-mm glass coverslips in 24-well plates. Cells were washed once in PBS before being fixed with 100% methanol for 10 min at room temperature. After three washes with PBS, the cells were incubated with FITC-conjugated heparan sulfate-specific monoclonal antibody (Seikagaku America, Ijamsville, MD) and diluted 1:1,250 for 1 h in the dark at room temperature. The monolayers were washed three times in PBS before being viewed and photographed.
Gene characterization.
Genomic DNA (5 μg) isolated from CHO-22 and CHO-18.4 cells was digested with HindIII restriction endonuclease (Roche Diagnostics, Indianapolis, IN), separated by electrophoresis on 0.5% agarose gels, and transferred to positively charged nylon membranes (Roche) for Southern hybridization or recovered for cloning. The nylon membrane was probed with a digoxigenin-labeled DNA fragment generated by PCR using primers to the neomycin resistance gene sequence (NeoF, 5′-GCACAACAGACAATCGGCTGC-3′; NeoR, 5′-CTTCAGCAATATCACGGGTAGC-3′) and using the pExchange module EC-Neo plasmid (Stratagene, La Jolla, CA) as the template. The membrane was hybridized overnight at 68°C and was washed at high stringency. The antidigoxigenin antibody was used to detect labeled DNA hybridized to the nylon membrane by a luminescent detection assay (Roche), and the film (Hyperfilm ECL; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) was exposed for 2.5 h at room temperature before being developed.
HindIII restriction endonuclease DNA fragments of approximately 7 kb to 8 kb were isolated from agarose gels and ligated into the pBluescript II phagemid vector and transformed into XL1-Blue MRF cells (Stratagene). The entire 1-ml transformation reaction mixtures were plated on nylon filters (Roche), placed on top of Luria agar plates containing 50 μg/ml ampicillin (Sigma), and incubated overnight at 37°C. Replica filters were made from each master filter and were prepared for hybridization. Approximately 200,000 total colonies were screened by hybridization with the digoxigenin-labeled fragment from the neomycin resistance gene described above. The filters were hybridized at 65°C overnight, washed, developed, and exposed to film. Positive clones were isolated and rehybridized with the labeled probe. Two independently positive clones from the original plates were isolated. Individual colonies were grown in 3-ml LB broth cultures supplemented with 50 μg/ml ampicillin overnight at 37°C. Plasmid DNA was isolated (QIAGEN, Valencia, CA), and the DNA sequence was obtained for the cloned fragment (Biomolecular Resource Center, University of California, San Francisco).
Total RNA was extracted from CHO-22, CHO-18.4, and L929 cells using RNeasy (QIAGEN). Approximately 2 μg of total RNA was added to the cDNA synthesis step in reverse transcriptase (RT) PCR using the ThermoScript RT-PCR system (Invitrogen, Carlsbad, CA). A primer specific to the Smd3 gene (Smd3RTR, 5′-GAGGACCTTGTCAAGCCACTGCAA-3′) was used to prime the cDNA synthesis reaction mixtures that were incubated at 50°C for 45 min. An additional primer was combined with SmdRTR (SmdRTF, 5′CAGAACGTCAACACCAAGTGC-3′) for the PCRs.
RESULTS
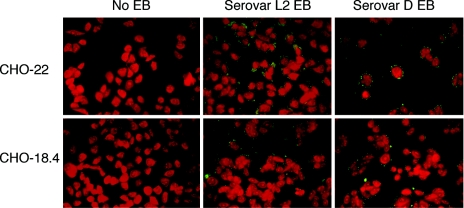
Immunofluorescence detection of C. trachomatis serovar L2 and serovar D was used to test the binding of EB to heparan sulfate-deficient CHO-18.4 cells. Both serovars bound the parental cell line CHO-22 (and CHO-K1, the cell line from which CHO-22 was derived; data not shown) and the mutant cell line CHO-18.4 in qualitatively similar amounts (Fig. 1). A large reduction in EB binding to CHO-18.4 cells was not observed, suggesting that the specific lack of heparan sulfate on the surface of CHO cells had little effect on the ability of EB to attach to host cells.
FIG. 1.
Cell attachment of C. trachomatis to CHO-22 and CHO-18.4 cells was tested in triplicate. Following attachment, chlamydial EB were visualized by staining with an FITC-conjugated monoclonal antibody. Similar data were obtained in at least three independent experiments for C. trachomatis (serovars L2 and D).
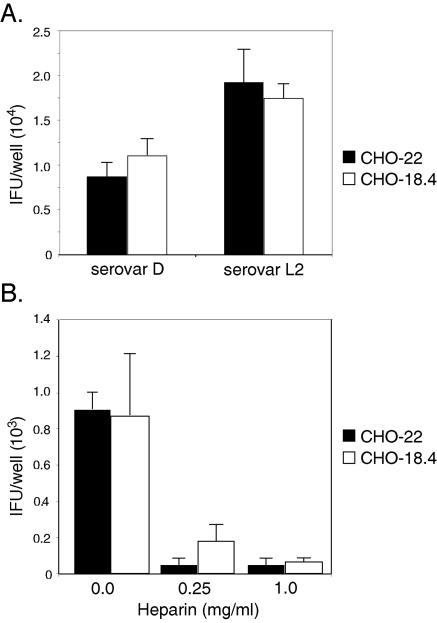
It was next tested whether there were differences in susceptibility to infection between these cell lines, as infectivity measurements reflect the sum of many steps, including attachment, and are sensitive and very specific. The infectivities of C. trachomatis serovar L2 and serovar D (representing two C. trachomatis biovariants) for CHO-22 and CHO-18.4 cells were not significantly different (Fig. 2A). Infectivity was tested over a range of EB concentrations, and in each case, there was not a difference (data not shown). The sensitivity to infection of these cell lines was also compared to that of CHO-K1, and no differences were observed (data not shown). These data were consistent with the attachment data and demonstrated that CHO-18.4 was as susceptible to infection by chlamydiae as was either CHO-K1 or CHO-22.
FIG. 2.
(A) Infectivity of C. trachomatis (serovar L2) and C. trachomatis (serovar D) for CHO-22 (black bars) and CHO-18.4 (white bars) cells. (B) Dose-dependent inhibition of infectivity for CHO-22 (black bars) and CHO-18.4 (white bars) cells by 0.25 or 1.0 mg/ml of heparin added at the time of tissue culture inoculation of organisms. At approximately 48 h postinfection, chlamydial inclusions were detected by immunofluorescence and enumerated by counting inclusions (IFU). Similar data were obtained in at least three independent experiments.
In vitro infection of host cells by C. trachomatis is >90% inhibited by the addition of exogenous heparin (4, 15, 26). If the basis of heparin inhibition is competition with host cell surface heparan sulfate, then CHO-18.4 cells will be resistant to heparin inhibition because they lack cell surface heparan sulfate. Inconsistent with this conclusion, the infectivity of CHO-22 or CHO-18.4 cells by both biovars of C. trachomatis was inhibited by the addition of similar amounts of exogenous heparin (Fig. 2B).
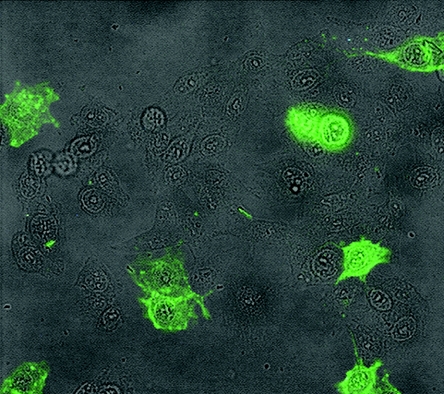
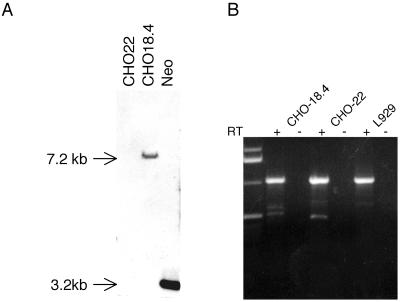
In the studies by Jan et al. (10), the CHO-18.4 cell line was shown to be deficient in heparan sulfate proteoglycans and to have a single retroviral insertion in an ∼7.2-kbp HindIII endonuclease restriction fragment, but the disrupted gene was unknown. We confirmed the biochemical tests for heparan sulfate deficiency by testing whether CHO-18.4 cells would bind a heparan sulfate-specific monoclonal antibody. By direct immunofluorescence, CHO-22 cells stained brightly, whereas CHO-18.4 cells did not stain with this antibody, demonstrating that CHO-18.4 cell surfaces lack heparan sulfate-containing proteoglycans (Fig. 3). We also confirmed that the CHO-18.4 cell line has a single retroviral insertion in an ∼7.2-kbp HindIII fragment by hybridization of genomic DNA with a labeled probe for the neomycin resistance gene (Fig. 4A). Moreover, we cloned the ∼7.2-kbp HindIII fragment containing the retroviral insertion and sequenced the flanking DNA. The 4,168 bp of CHO cell nonvector sequence did not reveal a similar sequence in GenBank by BLAST searching; however, BLAST searching of the mouse genome database revealed sequence similarity and localized the retroviral vector insertion within the first intron of murine SMAD3 (11) on chromosome 9. Primers were designed based upon the mouse genome sequence, and murine L929, CHO-22, and CHO-18.4 cells each produced similar-sized mRNA products by RT-PCR (Fig. 4B), suggesting that the retroviral insertion did not affect transcription of the hamster SMAD3. Although this analysis did not reveal which gene was affected by the promoter trap retroviral vector or rule out whether a fortuitous mutation occurred during the retroviral insertion and selection, the cell line is nevertheless deficient in cell surface proteoglycans that display heparan sulfate.
FIG. 3.
Immunofluorescence of CHO-22 and CHO-18.4 cells by a heparan sulfate-specific monoclonal antibody. CHO-22 and CHO-18.4 cells were mixed in culture at a ratio of approximately 1:9, fixed, and stained with FITC-conjugated heparan sulfate-specific monoclonal antibody. Unstained cells can be seen among those cells stained with antibody. Staining each cell line separately with FITC-conjugated heparan sulfate-specific monoclonal antibody resulted in no detectable staining for CHO-18.4 cells and uniformly positive staining for CHO-22 cells (data not shown).
FIG. 4.
(A) Southern hybridization of CHO-22 and CHO-18.4 cell genomic DNA digested with HindIII and probed with a labeled DNA fragment containing the neomycin resistance gene. (B) RT-PCR (with [+] or without [−] RT) of SMAD3 mRNA from CHO-18.4, CHO-22, and murine L929 cells.
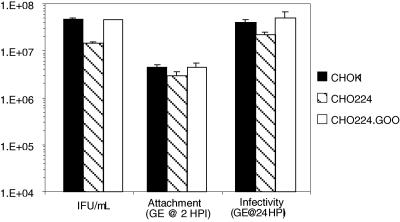
Because we could not associate a specific gene defect in the CHO-18.4 cells to enable restoration by complementation to its wild-type phenotype, we tested an alternative mutant CHO cell line that had previously been shown to be deficient in heparan sulfate, and its phenotype was restored by complementation of the gene defect. Bai et al. (1) identified the gene responsible for a heparan sulfate-deficient phenotype for the CHO-224 pgsG complementation group. The loss of glucuronosyltransferase I activity accounted for the PgsG phenotype as heparan sulfate biosynthesis was restored in CHO-224.GOO by stable complementation and expression of glucuronosyltransferase I cDNA. C. trachomatis infectivities differed twofold for CHO-K1, the heparan sulfate-deficient CHO-224, and its complemented strain CHO-224.GOO based on numbers of IFU determined by immunofluoresence (Fig. 5). Using quantitative real-time PCR, we tested both attachment and growth for C. trachomatis serovar L2 in these cell lines. Again, there was approximately a twofold reduction in both binding and infectivity in the mutant cell line lacking heparan sulfate (Fig. 5). These data show that there can be a small effect attributable to the presence of glucuronosyltransferase I expression; however, this does not approach a magnitude that can be related to the quantitative ablation of infectivity by competitive inhibition with exogenous heparan sulfate.
FIG. 5.
Attachment and infectivity of C. trachomatis (serovar L2) for CHO-K1, heparan sulfate-deficient CHO-224, and glucuronosyltransferase I-complemented CHO-224.GOO. At approximately 24 h postinfection, chlamydial inclusions were detected by immunofluorescence and enumerated by counting inclusions (IFU). Similar data were obtained in at least three independent experiments. Real-time quantitative PCR was used to determine the numbers of EB genome equivalents (GE) in wells containing cell monolayers after 2 h postinfection (HPI) at room temperature and washing, representing the number of chlamydiae attached to the monolayer (Attachment), and at 24 HPI, representing the amount of chlamydial growth (Infectivity).
DISCUSSION
While heparan sulfate-containing glycosaminoglycans on the surfaces of mammalian cells play important roles in microbial pathogen interactions (17, 19), the interpretation of data for infectivity by chlamydiae has been inconsistent. Chlamydiae are capable of binding heparan sulfate, and, consequently, the presence of heparan sulfate on the surfaces of host cells might be expected to affect adherence (2, 5, 12, 26). This interaction is further complicated by data from several laboratories showing that chlamydial EB display heparan sulfate or a functionally analogous compound on their surfaces prior to infection (2, 4, 8, 18, 25, 26). Moreover, it has recently been shown that fibronectin is also sequestered to the surfaces of EB as a natural outcome of in vitro infection and that fibronectin binding to EB is mediated by heparan sulfate lyase-sensitive components (13).
A popular and experimentally simple assessment has been to test the relative sensitivity to infection of CHO mutant cells deficient in the synthesis of heparan sulfate (19). Despite the straightforward experimental method, these data require circumspect judgement and interpretation for chlamydial infection. CHO-677, CHO-745, and CHO-761 cells are unequivocally deficient in the production of heparan sulfate, as shown by biochemical analysis and by complementation studies (1, 6, 7, 16). Each report to date has shown that CHO-677, CHO-745, and CHO-761 cells are less sensitive than the parental CHO-K1 to infection by C. trachomatis and C. pneumoniae by factors ranging from two- to ninefold. Several investigators have proposed that host cell surface heparan sulfate is either essential for chlamydial infection (23) or is the host cell receptor for chlamydia (20, 21, 24).
If cell surface heparan sulfate plays an essential or receptor role in chlamydial infection, then mutant cell lines deficient in heparan sulfate biosynthesis must be meaningfully less susceptible to infection by chlamydiae. The minor (e.g., ca. twofold) differences in infectivity between CHO-K1 and CHO-761 cells are inconsistent with this hypothesis. Taraktchoglou et al. (21) dismissed these data, claiming, without experimentation, that CHO-761 produces 5% wild-type heparan sulfate and that this accounts for its susceptibility to chlamydial infection. Comprehensive characterization of CHO-761 has shown that it is as deficient in heparan and chondroitin sulfate biosynthesis as is CHO-745 (1, 7). It has previously been shown that sulfated oligosaccharides are required for competitive inhibition of chlamydial infectivity (5); however, consistent with the lack of a functional role for host cell glycosaminoglycans, Yabushita et al. (24) found no inhibition of chlamydial infectivity for a mutant cell line deficient in sulfated glycosaminoglycans.
We tested the hypothesis that host cell surface heparan sulfate is required for chlamydial infection, but the test of this hypothesis failed, demonstrating that cell surface heparan sulfate does not serve an integral function in promoting chlamydial infection of mammalian cells. Exogenous heparin inhibited infection of CHO-18.4 cells despite the fact that these cells lack cell surface heparan sulfate. Thus, the molecular basis for the ability of heparin to ablate infection is not competition with EB for binding host cell surface heparan sulfate; rather, this finding is consistent with the hypothesis that heparin is competitively inhibiting a functionally analogous component on the surfaces of EB (26).
With deference to some previous interpretations (20, 21, 23, 24), minor reductions in chlamydial attachment or infectivity for CHO-677, CHO-745, or CHO-761 mutant cells are insufficient to warrant a conclusion that host cell heparan sulfate serves as a receptor for chlamydiae. These reproducible reductions in infectivity appear to be an epiphenomenon of chemical mutagenesis unrelated to the mutant cell's lack of glycosaminoglycan production.
Acknowledgments
We thank Laura Cieslik, Erin Banta, and Christina Shields for their excellent technical contributions to this research. We are very grateful to D. E. Griffin (Johns Hopkins University) for providing the CHO-22 and CHO-18.4 mutant cell lines and to J. Esko (University of California, San Diego) for contributing the CHO psgG mutant cell lines. We thank C. Fenner for editing the manuscript and critical discussions.
This research was supported by National Institutes of Health grants AI42156 and AI32943.
Editor: J. N. Weiser
REFERENCES
- 1.Bai, X., G. Wei, A. Sinha, and J. D. Esko. 1999. Chinese hamster ovary cell mutants defective in glycosaminoglycan assembly and glucuronosyltransferase I. J. Biol. Chem. 274:13017-13024. [DOI] [PubMed] [Google Scholar]
- 2.Beswick, E. J., A. Travelstead, and M. D. Cooper. 2003. Comparative studies of glycosaminoglycan involvement in Chlamydia pneumoniae and C. trachomatis invasion of host cells. J. Infect. Dis. 187:1291-1300. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J. C., and R. S. Stephens. 1997. Chlamydia trachomatis glycosaminoglycan-dependent and independent attachment to eukaryotic cells. Microb. Pathog. 22:23-30. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J. C., and R. S. Stephens. 1994. Trachoma and LGV biovars of Chlamydia trachomatis share the same glycosaminoglycan-dependent mechanism for infection of eukaryotic cells. Mol. Microbiol. 11:501-507. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. C., J. P. Zhang, and R. S. Stephens. 1996. Structural requirements of heparin binding to Chlamydia trachomatis. J. Biol. Chem. 271:11134-11140. [PubMed] [Google Scholar]
- 6.Esko, J. D., T. E. Stewart, and W. H. Taylor. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA 82:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esko, J. D., J. L. Weinke, W. H. Taylor, G. Ekborg, L. Roden, G. Anantharamaiah, and A. Gawish. 1987. Inhibition of chondroitin and heparan sulfate biosynthesis in Chinese hamster ovary cell mutants defective in galactosyltransferase I. J. Biol. Chem. 262:12189-12195. [PubMed] [Google Scholar]
- 8.Gutierrez-Martin, C. B., D. M. Ojcius, R. C. Hsia, R. Hellio, P. M. Baviol, and A. Dautry-Varsat. 1997. Heparin-mediated inhibition of Chlamydia psittaci adherence to HeLa cells. Microb. Pathog. 22:47-57. [DOI] [PubMed] [Google Scholar]
- 9.Huang, J., F. J. DeGraves, D. Gao, P. Feng, T. Schlapp, and B. Kaltenboeck. 2001. Quantitative detection of Chlamydia spp. by fluorescent PCR in the LightCycler. BioTechniques 30:150-157. [DOI] [PubMed] [Google Scholar]
- 10.Jan, J. T., A. P. Byrnes, and D. E. Griffin. 1999. Characterization of a Chinese hamster ovary cell line developed by retroviral insertional mutagenesis that is resistant to Sindbis virus infection. J. Virol. 73:4919-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kano, K., A. Notani, S. Y. Nam, M. Fujisawa, M. Kurohmaru, and Y. Hayashi. 1999. Cloning and studies of the mouse cDNA encoding Smad3. J. Vet. Med. Sci. 61:213-219. [DOI] [PubMed] [Google Scholar]
- 12.Kihlstrom, E., M. Majeed, B. Rozalska, and T. Wadstrom. 1992. Binding of Chlamydia trachomatis serovar L2 to collagen types I and IV, fibronectin, heparan sulphate, laminin and vitronectin. Zentbl. Bakteriol. 277:329-333. [DOI] [PubMed] [Google Scholar]
- 13.Kleba, B. J., E. Banta, E. A. Lindquist, and R. S. Stephens. 2002. Recruitment of mammalian cell fibronectin to the surface of Chlamydia trachomatis. Infect. Immun. 70:3935-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koehler, J. E., R. R. Burgess, N. E. Thompson, and R. S. Stephens. 1990. Chlamydia trachomatis RNA polymerase major σ subunit. Sequence and structural comparison of conserved and unique regions with Escherichia coli σ70 and Bacillus subtilis σ43. J. Biol. Chem. 265:13206-13214. [PubMed] [Google Scholar]
- 15.Kuo, C. C., and T. Grayston. 1976. Interaction of Chlamydia trachomatis organisms and HeLa 229 cells. Infect. Immun. 13:1103-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lidholt, K., J. L. Weinke, C. S. Kiser, F. N. Lugemwa, K. J. Bame, S. Cheifetz, J. Massaguāe, U. Lindahl, and J. D. Esko. 1992. A single mutation affects both N-acetylglucosaminyltransferase and glucuronosyltransferase activities in a Chinese hamster ovary cell mutant defective in heparan sulfate biosynthesis. Proc. Natl. Acad. Sci. USA 89:2267-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menozzi, F. D., K. Pethe, P. Bifani, F. Soncin, M. J. Brennan, and C. Locht. 2002. Enhanced bacterial virulence through exploitation of host glycosaminoglycans. Mol. Microbiol. 43:1379-1386. [DOI] [PubMed] [Google Scholar]
- 18.Rasmussen-Lathrop, S. J., K. Koshiyama, N. Phillips, and R. S. Stephens. 2000. Chlamydia-dependent biosynthesis of a heparan sulphate-like compound in eukaryotic cells. Cell. Microbiol. 2:137-144. [DOI] [PubMed] [Google Scholar]
- 19.Rostand, K. S., and J. D. Esko. 1997. Microbial adherence to and invasion through proteoglycans. Infect. Immun. 65:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su, H., L. Raymond, D. D. Rockey, E. Fischer, T. Hackstadt, and H. D. Caldwell. 1996. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc. Natl. Acad. Sci. USA 93:11143-11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taraktchoglou, M., A. A. Pacey, J. E. Turnbull, and A. Eley. 2001. Infectivity of Chlamydia trachomatis serovar LGV but not E is dependent on host cell heparan sulfate. Infect. Immun. 69:968-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Melchner, H., and H. E. Ruley. 1989. Identification of cellular promoters by using a retrovirus promoter trap. J. Virol. 63:3227-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wuppermann, F. N., J. H. Hegemann, and C. A. Jantos. 2001. Heparan sulfate-like glycosaminoglycan is a cellular receptor for Chlamydia pneumoniae. J. Infect. Dis. 184:181-187. [DOI] [PubMed] [Google Scholar]
- 24.Yabushita, H., Y. Noguchi, H. Habuchi, S. Ashikari, K. Nakabe, M. Fujita, M. Noguchi, J. D. Esko, and K. Kimata. 2002. Effects of chemically modified heparin on Chlamydia trachomatis serovar L2 infection of eukaryotic cells in culture. Glycobiology 12:345-351. [DOI] [PubMed] [Google Scholar]
- 25.Zaretzky, F. R., R. Pearce-Pratt, and D. M. Phillips. 1995. Sulfated polyanions block Chlamydia trachomatis infection of cervix-derived human epithelia. Infect. Immun. 63:3520-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, J. P., and R. S. Stephens. 1992. Mechanism of C. trachomatis attachment to eukaryotic host cells. Cell 69:861-869. [DOI] [PubMed] [Google Scholar]