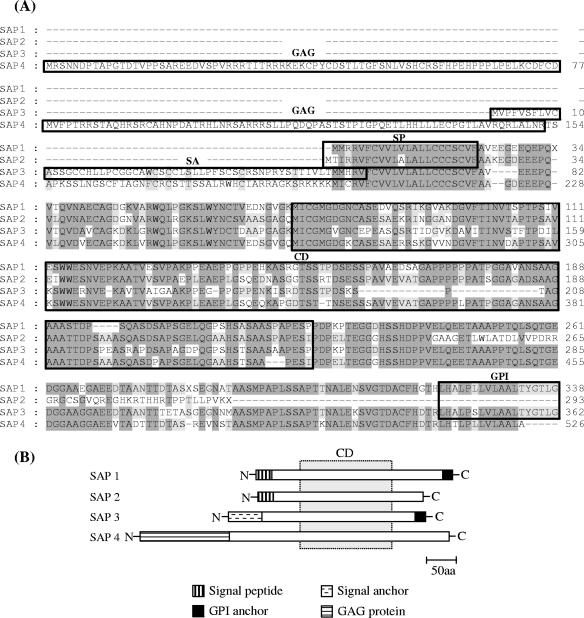
FIG. 2.
Multiple alignment of amino acid sequences from SAP variants. (A) Alignments were done by Clustal W with the MegAlign program (DNAstar Inc.). Dashes were introduced to maximize the alignment. Sequences corresponding to the conserved residues are shaded in dark gray (>80% conservation) and light gray (60 to 80% conservation), and the absence of shading denotes residues with <60% conservation. The N-terminal signal peptide (SP), the N-terminal signal anchor sequence (SA), the CD of approximately 155 amino acids common to all SAPs, and the C-terminal GPI anchor addition site are boxed. The N-terminal domain of the T. cruzi gag protein encoded by SAP 4 is also boxed. (B) Schematic drawing of SAPs indicating the regions of identity between different members of the SAP family. The dotted rectangle denotes the CD common to all SAPs.