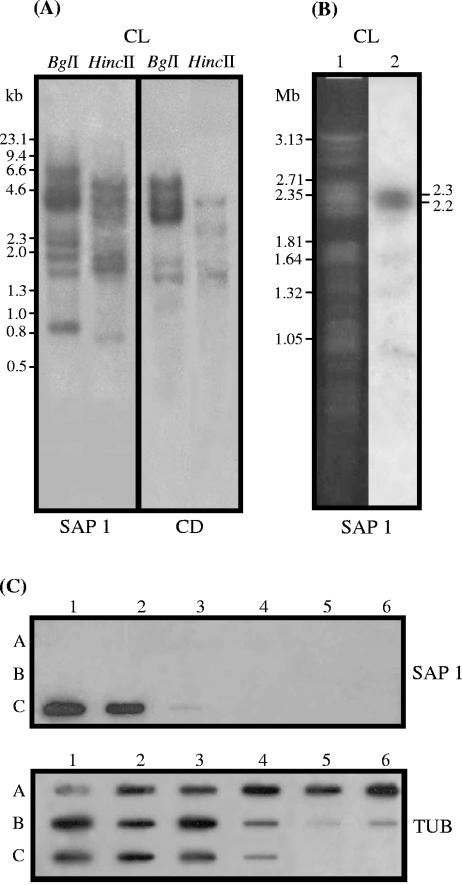
FIG. 4.
Genomic organization of SAP-related sequences. (A) Southern blot of T. cruzi (clone CL Brener) genomic DNA digested with BglI or HincII and hybridized with the 32P-labeled insert of SAP 1 or the fragment encoding the central domain of approximately 155 amino acids common to all SAPs. Numbers correspond to the molecular sizes in kilobases. (B) Chromosomal mapping of SAP-related sequences. Chromosomal bands of epimastigotes (clone CL Brener) were separated by pulsed-field gel electrophoresis, stained with ethidium bromide (lane 1), transferred onto nylon membranes, and hybridized with the 32P-labeled insert of SAP 1 (lane 2). Numbers correspond to the molecular sizes in megabases. (C) Demonstration of species specificity of SAP genes by slot blot hybridization using genomic DNA from different species from the genus Trypanosoma. (Rows A) Lanes: 1, T. rangeli (Saimiri); 2, T. rangeli (Preguiça); 3, T. rangeli (P.G.); 4, T. rangeli (Palma-2); 5, T. rangeli (Choachi); 6, T. rangeli (San Augustin). (Rows B) Lanes: 1, T. lewisi; 2, T. blanchardi; 3, T. rabinowitschae; 4, T. conorhini; 5, T. cyclops; 6, T. evansi. (Rows C) Lanes: 1, T. cruzi (G); 2, T. cruzi (Y); 3, T. dionisii; 4, Trypanosoma sp. strain 60. Samples were hybridized with random primed 32P-labeled SAP probe. After exposition, the membranes were incubated at 100°C for 30 min in 0.1% SDS and rehybridized with the tubulin gene probe.