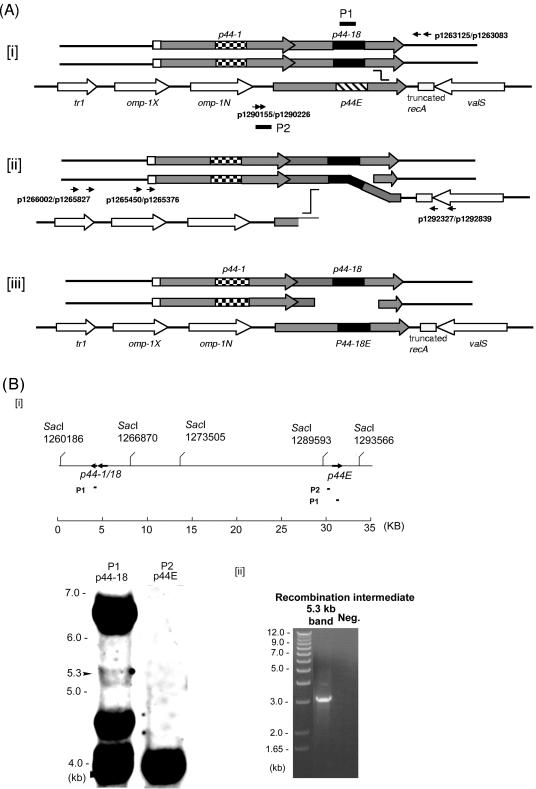
FIG. 1.
Analysis of the p44 recombination intermediate in A. phagocytophilum. (A) Successive half-crossover model for p44-18 conversion to the p44E and the experimental design. [i] The donor locus was expected to be a part of replicated chromosome; the p44-1/18 locus in a donor chromosome (p44-1/18D) synapses with the 3′ conserved region of p44E in the recipient chromosome. [ii] A half crossover between p44E and p44-1/18D generates one recombination duplex and two ends. The upstream of the synapsed region of p44E is presumably degraded to generate a single-stranded tail at the 5′ conserved region by an exonuclease. [iii] The putative final products of the successive crossover are one duplex with two double strands exchanged in the recipient p44 expression locus and two ends in the donor locus. The gray boxes are p44 conserved regions, and the large checkerboard, black, and wide downward diagonal are hypervariable regions of p44-1, p44-18, and the recipient p44E, respectively. The arrows of boxes indicate transcriptional orientation. Horizontal arrows with numbers indicate primers used to isolate the intermediate structure by PCR and primers used to demonstrate the absence of a reciprocal recombination product. Black short bars indicate the locations of two probes (P1 and P2) used for Southern blot analysis in Fig. 1B. The “stairstep” symbol indicates the potential pair of the recombination sites. (B) Southern blot analysis of the half-crossover intermediate structure of A. phagocytophilum cultured in HL-60 cells. [i] The probe positions and SacI digestion sites within the 35-kb region containing p44-1/18 and p44E are indicated. The numbers under the SacI sites indicate the positions of the cleavage sites in the genome. The genomic DNA was purified from A. phagocytophilum cultivated in HL-60 cells. Southern blot analysis of genomic DNA digested with the restriction enzyme SacI with two probes (P1 and P2) showed the ∼6.6-kb band containing the donor p44-1/18 locus and a 4-kb band containing the recipient p44E. An ∼5.3-kb band contained duplicated p44-1/18. [ii] PCR was performed using the primer pairs p1265450-p1292839 and p1265376-p1292327 (Fig. 1Aii) and the DNA isolated from the 5.3-kb region of the gel as a template. The numbers on the left indicate the molecular sizes. A band of approximately 3.2 kb was amplified and sequenced GenBank accession no. DQ011270. “Neg.” refers to a negative control with water as a template.