Abstract
This study describes the presence of 10 hemolysin orthologs in the genome of the opportunistic human anaerobic pathogen Bacteroides fragilis, which is currently classified as a nonhemolytic bacterium. The hemolysins were designated HlyA through HlyI plus HlyIII. All cloned hemolysin genes were able to confer hemolytic activity to a nonhemolytic Escherichia coli strain on blood agar plates. Interestingly, HlyH was found to be present in the genome of the B. fragilis NCTC9343 strain but absent in strains 638R, YCH46, and Bacteroides thetaiotaomicron VPI-5482. The hemolysins HlyA, HlyB, and HlyIII were selected for further characterization. HlyA, HlyB, and HlyIII were cytolytic to erythrocytes on liquid hemolytic assay. When hlyA and hlyB were expressed together in a nonhemolytic E. coli strain, the strain showed enhanced hemolytic activity on blood agar plates. Further analysis revealed that HlyA and HlyB have synergistic hemolytic activity as detected by the liquid hemolytic assay. In addition, the two-component hemolysins HlyA and HlyB form a protein-protein complex in vivo as determined by bacterial two-hybrid system assay. The hlyB and hlyA genes are organized in an operon that is coordinately regulated by iron and oxygen. Northern blot hybridization analysis revealed that hlyBA were expressed as a bicistronic mRNA induced approximately 2.5-fold under low-iron conditions and repressed in iron-rich medium. The normal iron-regulated expression of hlyBA mRNA was lost in the furA mutant strain. In contrast, the hlyA gene was also expressed as a single mRNA in iron-rich medium, but its expression was reduced approximately threefold under low-iron conditions in a Fur-independent manner. This suggests that hlyA alone is regulated by an unidentified iron-dependent regulator. Moreover, the expression levels of hlyBA and hlyA were reduced about threefold following oxygen exposure and treatment with hydrogen peroxide. Taken together, these results suggest that iron and oxidative stress have an effect on the control of hlyBA and hlyA transcriptional levels. A hlyBA mutant was constructed, and its hemolytic activity was greatly diminished compared to those of the hlyIII mutant and parent strains. In addition, the hlyBA mutant had a significant modification in colony morphology and growth deficiency compared to the parent strain. The implications of these findings for the pathophysiology of B. fragilis in extraintestinal infections and competition in ecological systems for this organism are discussed.
In the human colon (where at least 500 species of bacteria have been reported to reside), Bacteroides species account for about 30 to 40% of the total bacteria present in some people (20, 26, 43, 54). Bacteroides spp. are also important opportunistic human pathogens frequently isolated from anaerobic infections (11). Within the Bacteroides family, Bacteroides fragilis accounts for about 0.5% to 1% of total Bacteroides found in the human large intestinal tract (11). Despite its low frequency as a component of the intestinal microflora, B. fragilis is by far the number one anaerobe isolated from anaerobic infections. It accounts for 50 to 70% of all anaerobes isolated from human infections, such as intra-abdominal infections, infections of the female genital tract, deep wounds, brain abscesses, and bacteremia (4, 5, 11).
Although the whole arsenal of B. fragilis virulence determinants is unclear, certain factors, such as capsular polysaccharides, microbial adherence, production of proteases and neuraminidase, and inhibition of phagocytosis, are considered important (43). The most studied B. fragilis virulence factor associated with pathogenicity is the production of eight distinct capsular polysaccharide complexes and their relation to abscess formation (14, 15, 21). Though adherence, lipopolysaccharide, and the production of neuraminidase, enterotoxin, and proteolytic enzymes might play a role in B. fragilis pathogenicity, they are not currently recognized to be strong determinants of virulence (43).
There are other factors that are considered important in infections by aerobes and facultative bacteria, but they have been given little attention as far as their role in the pathogenesis of Bacteroides spp. is concerned. One such factor is the ability of B. fragilis to produce hemolysins or cytolysins. Hemolysins have been reported to be powerful virulence determinants in both gram-positive and gram-negative bacteria (24, 38, 51). Hemolysins are cytotoxic proteins that target cell membranes, and their mechanisms of damaging membrane integrity can be classified into three major groups: enzymatic activity, pore-forming cytolysin, or surfactant (51). Many microbial hemolysins offer an advantage to the bacteria by lysing and killing incoming leukocytes, thus promoting survival of the microbe by not only weakening the immune system but also by gaining access to nutrients (24). Moreover, bacterial cytolysins/hemolysins play an important role in the equilibrium control of the microbial ecosystem population associated with eukaryotic hosts (9).
To date, no hemolysin has been characterized in B. fragilis despite the fact that rare strains have a hemolytic phenotype (18, 19). Nevertheless, a search of the B. fragilis genome sequence available at http://www.sanger.ac.uk/Projects/B_fragilis/revealed the presence of 10 genes with homology to other bacterial hemolysins (K. P. Robertson, C. J. Smith, and E. R. Rocha, Abstr. 104th ASM Gen. Meet. Am. Soc. Microbiol. 2004, abstr. B-042, 2004). These findings were intriguing because this anaerobic pathogen is classified as a nonhemolytic bacterium (18, 19, 53). To clarify this apparent contradiction, putative hemolysin genes found in the B. fragilis genome were cloned and found to confer a hemolytic phenotype in a nonhemolytic Escherichia coli strain (K. P. Robertson, C. J. Smith, and E. R. Rocha, Abstr. Anaerobe 7th Biennial Cong. Anaer. Soc. Am., abstr. II-O2, 2004). More recently, independent studies have confirmed our observation that there is an extensive number of putative hemolysin homologues in the annotated genome sequences of B. fragilis NCTC9343 and YCH46 (7, 22). Interestingly, this extensive number of putative hemolysin genes does not correlate with the “nonhemolytic” characteristic phenotype of most clinical and laboratory strains of B. fragilis (18, 19, 53). Therefore, these findings prompted us to characterize these putative hemolysin genes in order to confirm their hemolytic/cytolytic properties and role in B. fragilis pathophysiology. It is important to mention that the only other cytotoxin that has been studied in B. fragilis, fragilysin, is a Zn-metalloprotease enterotoxin that is cytotoxic only to the human intestinal carcinoma cell line HT29/C1. It specifically cleaves E-cadherin of the zone adherens (40). Moreover, fragilysin is not membranolytic to eukaryotic cells (50), nor does it seem to be a virulence factor for extraintestinal infections (12). These properties are much different than the characteristics of hemolysins mentioned above.
In this study we report the characterization of B. fragilis hemolysins HlyA, HlyB, and HlyIII. We also show that HlyA and HlyB are two-component cytolysins that act together to enhance their respective singular hemolytic activities against erythrocytes. The hlyB and hlyA genes are organized in an operon whose expression is differentially regulated by oxygen and iron availability. In addition, we show that a hlyBA mutant has a diminished hemolytic activity, altered colony morphology, and a growth defect in vitro.
MATERIALS AND METHODS
Strains, media, and growth conditions.
B. fragilis strains used in this study are listed in Table 1. B. fragilis strain NCTC9343 is isogenic to ATCC 25285, and henceforth ATCC 25285 was used in this study for experimental purposes when required. Strains were routinely grown on BHIS (brain heart infusion supplemented with l-cysteine, hemin, and NaHCO3) agar. Rifamycin (20 μg/ml), 100 μg/ml gentamicin, 5 μg/ml tetracycline, 10 μg/ml erythromycin, and 25 μg/ml cefoxitin were added to the media when required. BHIS-blood agar plate (BAP) is BHIS agar supplemented with 5% defibrinated blood from sheep, horses, rabbits, cows (Gemini Bio-Products, Woodland, CA), or adult human volunteers. For some experiments, bacteria were grown on a modified defined medium supplemented with 5% defibrinated blood (DM-BAP) and on a semidefined medium as previously described (33). E. coli strains were inoculated in Luria-Bertani agar (L-agar) supplemented with 40 μg/ml isopropyl-ß-d-thiogalactopyranoside (IPTG) and 100 μg/ml ampicillin. Human, horse, rabbit, sheep, or bovine defibrinated blood (5%) (Gemini Bio-Products, Woodland, CA) was added when appropriate. Glucose (0.2%) was added to L-agar for anaerobic growth of E. coli.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotype or descriptiona | Source or referenceb |
|---|---|---|
| B. fragilis | ||
| ATCC 23745 | Clinical isolate, pleural fluid | ATCC |
| ATCC 25285 | Clinical isolate, appendix abscess (same as NCTC 9343) | ATCC |
| VPI 2393 | Unknown | VPI |
| 638R | Clinical isolate, Rifr | 30 |
| BER-2 | 638R ΔfurA Rifr Cfxr | This study |
| BER-41 | 638R ΔhlyBA::tetQ Rifr Tetr | This study |
| BER-45 | 638R ΔhlyIII::cfxA Rifr Cfxr | This study |
| BER-46 | 638R ΔhlyBA::tetQ ΔhlyIII::cfxA Rifr Tetr Cfxr | This study |
| E. coli | ||
| DH10B | Cloning host strain | Invitrogen |
| YMZ19 | MC1061 clyA::kan | 47 |
| Plasmids | ||
| pUC19 | Cloning vector, Ampr | Invitrogen |
| pGEM-T | Cloning vector | Promega |
| pFD516 | Suicide vector, derived from deletion of pBI143 in pFD288 (Tetr) (Spr) (Ermr) | 41 |
| pER-2 | 1,047-bp dsDNA fragment amplified from 638R chromosome containing hlyF ORF was cloned into the SphI/BamHI sites of pUC19 | This study |
| pER-3 | 1,445-bp dsDNA fragment amplified from 638R chromosome containing hlyE ORF was cloned into the SphI/BamHI sites of pUC19 | This study |
| pER-9 | 3.2-kb DNA fragment from 638R containing a 363-bp in-frame deletion in furA plus 2.1-kb cfxA gene cloned into the intergenic region cloned into pGEM-T | This study |
| pER-10 | 5.5-kb DNA fragment from pER-9 containing the ΔfurA construct was cloned into the BamHI/SstI sites of pFD516 | This study |
| pER-11 | 1,170-bp dsDNA fragment amplified from 638R chromosome containing hlyA ORF was cloned into the BamHI/SstI sites of pUC19 | This study |
| pER-12 | 933-bp dsDNA fragment amplified from 638R chromosome containing hlyB ORF was cloned into the BamHI/SstI blunted sites of pUC19 | This study |
| pER-13 | 1,477-bp dsDNA fragment amplified from 638R chromosome containing hlyG ORF was cloned into the BamHI/SstI sites of pUC19 | This study |
| pER-14 | 1,139-bp dsDNA fragment amplified from ATCC 25285 chromosome containing hlyH ORF was cloned into the BamHI/SstI sites of pUC19 | This study |
| pER-15 | 1,272-bp dsDNA fragment amplified from 638R chromosome containing hlyC ORF was cloned into the BamHI/SstI sites of pUC19 | This study |
| pER-16 | 363-bp dsDNA fragment amplified from 638R chromosome containing hlyD ORF was cloned into the BamHI/SstI sites of pUC19 | This study |
| pER-17 | 820-bp dsDNA fragment amplified from 638R chromosome containing hlyIII ORF was cloned into the BamHI/SstI sites of pUC19 | This study |
| pER-18 | 2,035-bp dsDNA fragment amplified from 638R chromosome containing hlyBA ORFs was cloned into the BamHI/SstI sites of pUC19 | This study |
| pER-61 | Suicide vector containing a ΔhlyBA construct in pFD516 | This study |
| pER-62 | 2.4-kb SstI fragment containing the tetQ gene cloned into the unique SstI site of pER-61 to make a ΔhlyBA::tetQ construct in pFD516 | This study |
| pER-64 | Suicide vector containing a ΔhlyIII::cfxA construct in pFD516 | This study |
Rifr, rifampin resistance; Cfxr, cefoxitine resistance; Tetr, tetracycline resistance; Ampr, ampicillin resistance; Spr, spectinomycin resistance; Ermr, erythromycin resistance. Antibiotic resistance expression in E. coli is indicated by drug resistance phenotypes shown in parentheses. dsDNA, double-stranded DNA.
ATCC, American Type Culture Collection (Manassas, VA); VPI, Anaerobe Laboratory, Virginia Polytechnic Institute and State University (Blacksburg, VA).
Cloning of hemolysin genes.
The hemolysin genes listed in Table 1 were amplified by PCR using oligonucleotide primers designed to amplify the entire open reading frame (ORF) plus an additional 15 to 20 nucleotides (nt) upstream of the translation start site. Restriction sites were incorporated in the oligonucleotide primers for direct cloning of the PCR products into the cloning vector pUC19 in the same orientation as lacZ. hlyA through hlyG and hlyIII were amplified from the B. fragilis 638R genome, while hlyH was amplified from the ATCC 25285 genome. Plasmids were transformed into nonhemolytic E. coli strains DH10B and screened for hemolytic activity on Luria-Bertani agar plates incubated at 37°C for 24 to 48 h. In some experiments E. coli YMZ19 was used as the host strain to confirm that the cloned genes were not interfering with endogenous E. coli hemolytic activity. Hemolytic activity was detected by the appearance of clear hemolytic zones around the bacterial colonies compared to the control host strain carrying vector alone.
Measurement of hemolytic activity.
Bacteria were harvested by centrifugation at 10,000 × g for 10 min. The cell pellet was washed with phosphate-buffered saline (PBS) (50 mM phosphate buffer, pH 7.4, 150 mM NaCl) and resuspended in 5 ml PBS. Cell lysates were obtained by using a French press. Whole cells and cell debris were separated at 10,000 × g for 30 min. Lysates were maintained at 4°C or stored at −70°C until needed. For liquid hemolytic assays, a protocol modified from the methods of Bernheimer (1) and Rowe and Welch (38) was used. Briefly, sheep red blood cells (RBCs) were washed with PBS and centrifuged at 400 × g for 5 min. Washes were repeated until the supernatant was visibly clear of hemoglobin. Erythrocyte suspensions were adjusted to 1% with PBS supplemented with 0.1% bovine serum albumin. Crude extracts were adjusted to approximately 2 mg/ml of protein. Then, 0.5 ml of this solution was incubated with 0.5 ml of the 1% erythrocyte suspension at 37°C. Samples were centrifuged at 120 × g for 7 min to remove undamaged RBCs. The concentration of released hemoglobin is estimated by reading absorbance at 545 nm in a spectrophotometer against a control background lysis solution (0.5 ml erythrocyte suspension with 0.5 ml PBS). A 100% hemolysis standard was obtained by mixing 1 volume of distilled water containing 0.04% saponin and 1 volume of 1% RBC suspension.
Construction of hlyBA and hlyIII deletion mutants.
A 1.43-kb chromosomal DNA fragment upstream from hlyB, including the first 57 nt within the N-terminal region, was amplified by PCR and cloned into the unique BamHI site of the E. coli-Bacteroides shuttle suicide vector pFD516 (42). Subsequently, a 1.68-kb DNA fragment downstream from hlyA, containing the last 155 nt of the hlyA C-terminal region, was amplified by PCR and cloned into the unique EcoRI site of the new construct pER-61. A 2.4-kb SstI fragment containing the tetracycline resistance gene tetQ was cloned into the unique SstI site of pER-61 to replace the internal 1.65-kb DNA fragment deleted from the hlyBA. The new plasmid pER-62, containing the ΔhlyBA::tetQ construct, was mobilized from E. coli DH10B into B. fragilis 638R by triparental filter mating protocols (41). Transconjugants were selected on BHIS agar containing 20 μg of rifamycin per ml, 100 μg of gentamicin per ml, and 5 μg of tetracycline per ml. Determination of sensitivity to either tetracycline or erythromycin was carried out to identify recombinants that were tetracycline resistant and erythromycin sensitive. Southern blot analysis was used to confirm the double-crossover genetic allele exchange of pER-62 into the B. fragilis chromosome. A transconjugant, BER-41, containing the ΔhlyBA::tetQ construct inserted into the B. fragilis 638R chromosome was selected for further studies.
To construct the hlyIII deletion mutant, a 520-bp internal DNA fragment from hlyIII was deleted and replaced by a 2.1-kb cefoxitin resistance gene, cfxA. Briefly, a 2.1-kb BamHI/EcoRI cfxA DNA fragment was cloned into the unique BamHI and EcoRI sites of the suicide vector pFD516. Then, a 1.52-kb DNA fragment upstream and a 1.60-kb DNA fragment downstream of the hlyIII gene were cloned, respectively, into the unique SphI/BamHI and EcoRI restriction sites of the pFD516/cfxA construct. The new plasmid, pER-64, carrying the ΔhlyIII::cfxA construct was mobilized into B. fragilis as described above. Transconjugants were selected on BHIS agar containing 20 μg of rifamycin per ml, 100 μg of gentamicin per ml, and 25 μg of cefoxitin per ml. Sensitivity to either cefoxitin or erythromycin was carried out to identify recombinants that were cefoxitin resistant and erythromycin sensitive as described above. A B. fragilis transconjugant, BER-45, containing the ΔhlyIII::cfxA construct inserted into the chromosome by double-crossover allelic recombination was selected for further analysis. The hlyBA::tetQ hlyIII::cfxA double mutant strain, BER-46, was obtained by mobilizing pER45 into strain BER-41 as described above.
Construction of a furA null mutant.
Briefly, a 275-bp EcoRV/NruI DNA fragment containing the first 25 N-terminal codons and 200 bp upstream of the furA promoter region was ligated in frame to a 1.7-kb NruI/SstI DNA fragment containing the last 17 furA codons and downstream region into pGEM-T (Promega, Madison, WI). The null mutation construct contained an in-frame deletion of 363 nt from the furA gene. Next, a 2.1-kb BamHI/blunted EcoRI cfxA gene was cloned into the BamHI/EcoRV sites of the new construct. Then, a 1.5-kb BamHI/BglII DNA fragment upstream of the furA promoter region was cloned into the BamHI site of the new construct pER-9. A 5.5-kb BamHI/SstI DNA fragment from pER-9 was cloned into the unique BamHI/SstI sites of the suicide vector pFD516. The new plasmid, pER10, was mobilized into B. fragilis 638R as described above. The mutant strain BER-2 containing the ΔfurA construct inserted into the chromosome by double-crossover allelic recombination was grown under iron-replete and iron-limiting conditions. Total RNA was extracted as described below. Real-time reverse transcription-PCR of feoAB transcripts and feoAB::xylB transcriptional fusion analysis were carried out to determine whether strain BER-2 lost the normal iron regulation uptake mechanism controlled by the ferric uptake regulator FurA in other bacteria (23). Both methods showed that feoAB mRNA expression was no longer repressed under high-iron conditions compared to the parent strain, confirming the mutation and loss of normal iron-responsive FurA regulation (data not shown).
Bacterial two-hybrid system assay.
hlyB was cloned into the “bait” vector pBT (BacterioMatch II; Stratagene, La Jolla, CA) in frame with λcI. hlyA was cloned into the “prey” vector pTRG in frame with the α-subunit of RNA polymerase according to the manufacturer's instructions. The new constructs were cotransformed into the E. coli two-hybrid system reporter strain derived from XL1-Blue MRF′ kan [Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn5 (Kanr)] containing the HIS3-aadA reporter cassette (Stratagene) and plated on selective screening medium. The selective screening plate is histidine-dropout M9 agar supplemented with 0.5 mM IPTG, 10 μg/ml tetracycline, 25 μg/ml chloramphenicol, and 5 mM 3-amino-1,2,4-triazole. Culture media were prepared according to the manufacturer's instructions. Transformants from selective medium were grown on dual-selective medium to confirm the interaction of the loaded “prey” and loaded “bait” constructs by the activation of the dual reporter system assay. Dual-selective screening plates are the same as selective medium but 12.5 μg/ml streptomycin is added. Self-activation controls are the E. coli two-hybrid system reporter strain carrying the following constructs: control 1, empty “bait” (pBT alone) cotransformed with loaded “prey” (pTRG/HlyA); control 2, loaded “bait” (pBT/HlyB) cotransformed with empty “prey” (pTRG alone).
RNA extraction and Northern blot hybridization.
Bacteria were grown in semidefined medium (SDM) as previously described (33). Hemin was replaced by 5 μg/ml protoporphyrin IX when required. Addition of 50 μM 2,2′-bipyridyl and 100 μM desferrioxamine was used to restrict iron availability in SDM. Ferrous sulfate at 100 μM was added as indicated in the text. For oxidative stress experiments, cultures were grown to an A550 of 0.3 and treated with 50 μM H2O2 for 5 min prior to total RNA extraction. To induce oxygen stress, cultures were split in half, one half was kept anaerobically and the other half was shaken aerobically at 250 rpm at a volume/flask ratio of 1/5 as previously described. Total RNA extraction and Northern blot analysis of mRNA were carried out as previously described (32), and internal fragments of hlyA and hlyB were used as specific probes. Densitometry analysis of the autoradiograph was normalized to the relative intensity of total 23S and 16S rRNA detected on the ethidium bromide-stained agarose gel to correct for any loading differences.
RESULTS
Characterization of B. fragilis hemolysins was initiated following the release of the B. fragilis NCTC9343 (ATCC 25285) and 638R genome sequences at http://www.sanger.ac.uk/Projects/B_fragilis/. A search in the NCTC9343 genome revealed the presence of 10 putative genes with homology to known bacterial hemolysin genes available in the GenBank database. Henceforth they are designated hlyA through hlyI plus hlyIII. Strain 638R has all the same hemolysin genes as NCTC9343 except for hlyH (Fig. 1 and data not shown). Moreover, hlyH was found to be part of a 25-kbp region found in the NCTC9343 strain but absent in strains 638R and YCH46. The hlyH chromosomal region is flanked by insertion sequence elements, and we speculate that it might be an unidentified putative pathogenicity island unique to NCTC9343 strain (unpublished data). The release of the complete genomic annotations of strains NCTC9343 (7) and YCH46 (22) confirmed our previous assumption that putative hemolysins are indeed present in the B. fragilis genome.
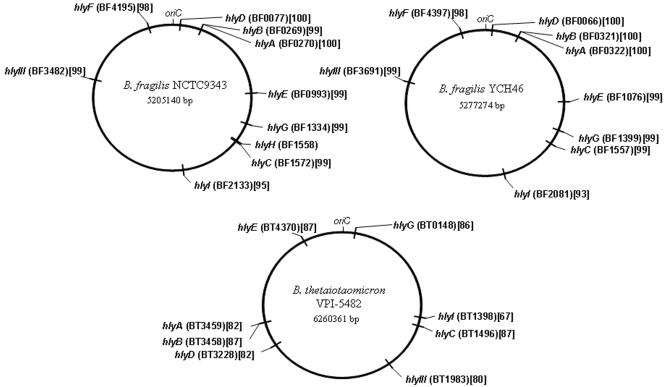
FIG. 1.
Circular map showing the chromosomal locations of putative hemolysins within B. fragilis and B. thetaiotaomicron strains. The peptide sequence for each hemolysin from strain 638R was used to identify the homologous peptide and respective locus tag number in B. fragilis NCTC9343 and YCH46 and B. thetaiotaomicron VPI-5482. The locus tags for strain 638R are not shown, because the genome annotation has not yet been released to public domain databases though the complete genome sequence is available at http://www.sanger.ac.uk/Projects/B_fragilis/. The locus tag locations shown in parentheses were retrieved from references 7, 22, and 55 and from websites and an FTP site (http://www.sanger.ac.uk/Projects/B_fragilis/, ftp://ftp.sanger.ac.uk/pub/pathogens/bf/, and http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=60491031). The percentage of amino acid identity of each gene product compared to that in strain 638R is shown in brackets. The genome length (in nucleotides) is depicted in each panel below the respective strain name.
A comparison of the genetic organization of the putative hemolysins in B. fragilis species revealed that hlyIII and hlyA through hlyI are located in similar positions in the chromosomes of strains NCTC9343, 638R, and YCH46 (Fig. 1 and data not shown). In contrast, the loci of hemolysin orthologs in Bacteroides thetaiotaomicron VPI-5482 (55) are found at different locations compared to B. fragilis relative to the chromosome origin of replication oriC (Fig. 1). This suggests that there have been major genetic rearrangements between these two species. Both hlyF and hlyH homologues are missing in the B. thetaiotaomicron VPI-5482 genome.
Hemolytic activity on BAP.
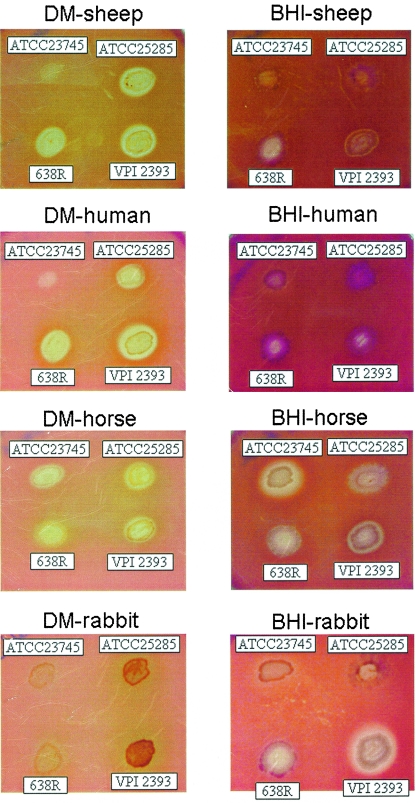
When strains ATCC 25285, 638R, ATCC 23745, and VPI 2393 were grown on DM-BAP, they produced beta-hemolysis after 3 to 5 days, but much weaker activity was observed on BHIS-BAP during the same period of incubation (Fig. 2). The early appearance of beta-hemolytic zones on DM-BAP compared to BHI-BAP was an indication that the production of hemolysin(s) might be under the control of a nutritional/starvation mechanism. These results suggest that the apparent lack of the hemolytic phenotype in clinical isolates of B. fragilis may be the consequence of a short period of incubation typically used in clinical laboratory diagnosis procedures. Strains ATCC 25285, 638R, and VPI 2393, were hemolytic for sheep, human, and horse red blood cells on DM-BAP. In contrast, strain ATCC 23745 showed strong hemolytic activity to horse RBCs but not to sheep and human RBCs (Fig. 2). Interestingly, no apparent hemolytic activity was detected on DM-BAP containing rabbit RBCs, although strains ATCC 27285 and VPI 2393 produced dark brown pigmentation. Furthermore, strains ATCC 23745 and VPI 2393 showed strong hemolytic activity against horse and rabbit RBCs on BHI-BAP. These findings suggest that the hemolytic activity produced by B. fragilis might be influenced by differences in strain background, the origin of the RBCs, and growth conditions.
FIG. 2.
Hemolytic activity of B. fragilis strains grown on defined medium (DM) and BHIS agar supplemented with 5% defibrinated blood. Plates were incubated anaerobically at 37°C for 5 days. Strain designations and animal blood source are shown in each panel.
Cloning of hemolysin genes.
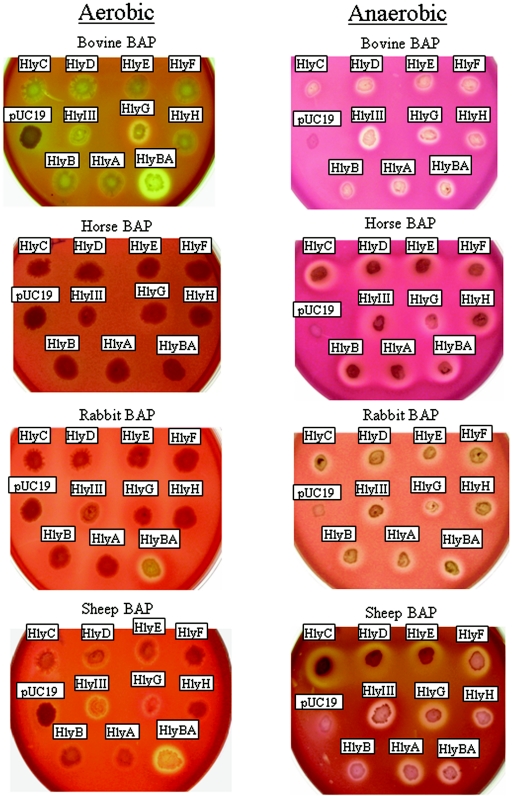
To further characterize B. fragilis hemolysins at a molecular level, hlyA through hlyH plus hlyIII were cloned, overexpressed in the host strain E. coli DH10B, and investigated for their ability to lyse RBCs. hlyI was not characterized in this study. Figure 3 shows that compared to controls with vector alone, all cloned genes conferred beta-hemolytic phenotype to E. coli grown under anaerobic conditions on BAP containing different animal RBCs. In contrast, there were differences in the hemolytic patterns when cells were grown under aerobic conditions. For example, horse RBCs did not show detectable hemolytic activity aerobically but were highly susceptible to hemolysis anaerobically. On bovine BAP, HlyG was beta-hemolytic under both aerobic and anaerobic conditions, while HlyIII showed alpha-hemolysis aerobically and stronger beta-hemolysis anaerobically. Interestingly, HlyB and HlyA showed alpha-hemolysis on bovine, rabbit, and sheep RBCs when expressed alone, but when expressed together, they showed ß-hemolysis, suggesting that they may act synergistically under aerobic conditions. In contrast, in the absence of oxygen, HlyB, HlyA, and HlyBA were beta-hemolytic. Taken together, these data clearly demonstrate that major differences exist in the activity of these hemolysins depending on the animal source of the RBCs and the presence or absence of oxygen.
FIG. 3.
Hemolytic activity of E. coli DH10B transformed with pUC19 carrying B. fragilis hemolysin genes. Bacteria were grown aerobically on L-agar medium supplemented with 5% defibrinated blood (BAP) and 0.5 mM IPTG. For anaerobic growth, media were supplemented with 0.2% glucose. E. coli carrying pUC19 alone was used as a control vector. All genes were amplified from B. fragilis 638R except for hlyH which was amplified from ATCC 25285 strain. Hemolytic activity was determined by the appearance of a clear zone around the growth (beta-hemolysis) or the appearance of a greenish hemolytic zone around the colonies (alpha-hemolysis).
Analysis of HlyA and HlyB hemolytic activity in the E. coli clyA mutant.
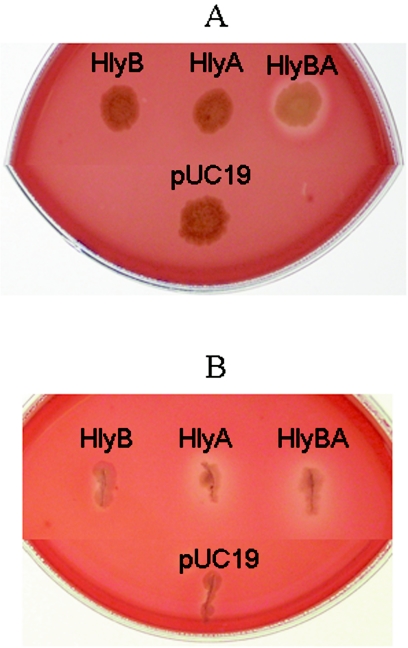
At this point of the investigation, we were particularly interested in further investigating HlyA and HlyB because our findings showed that they might act synergistically (Fig. 3). Plasmids pER-11, pER-12, and pER-18 were transformed into E. coli strain YMZ19 deleted for its cryptic, chromosome-encoded, silent hemolysin ClyA (49). When HlyB and HlyA were expressed together in pER-18, they showed synergistic hemolytic activity on the surface of human BAP incubated aerobically, but no activity when expressed alone compared to the vector control strain (Fig. 4). In contrast, in stabbed agar cultures, HlyBA and HlyA alone were hemolytic, while HlyB itself showed no detectable hemolysis under the same conditions compared to control vector. These findings confirm our hypotheses that HlyA and HlyB have synergistic interactions and that their activity was affected by exposure to air.
FIG. 4.
Hemolytic activity of E. coli YMZ19 (clyA::kan) expressing B. fragilis hemolysins HlyB, HlyA, and HlyBA and the vector pUC19 alone. Cultures were inoculated on L-agar plates containing 5% defibrinated human blood plus 0.5 mM IPTG, 100 μg/ml ampicillin, and 50 μg/ml kanamycin. (A) Bacteria inoculated on top of the agar. (B) Bacteria were stabbed through the blood agar.
Liquid hemolytic activity assay.
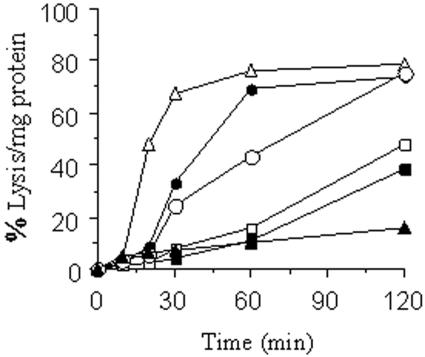
Cytotoxicity (measured by lysis of erythrocytes) of HlyBA, HlyB, HlyA, and HlyIII against sheep RBCs was determined by the liquid hemolytic assay (Fig. 5). Crude extracts of E. coli expressing HlyBA showed strong hemolytic activity against sheep RBCs, as 50% lysis occurred in just over 30 min of incubation. However, there was a weaker hemolytic activity in crude extracts containing HlyB or HlyA expressed alone. When crude extracts containing HlyB and HlyA were mixed together before addition to RBCs, hemolytic activity was restored similar to extracts containing HlyBA. These results correlate with previous data and add further support for the fact that HlyB and HlyA may have synergistic properties. Under the same conditions, HlyIII lysed 50% of RBCs in about 20 min at 37°C similar to lysis obtained with the HlyBA extract.
FIG. 5.
Liquid hemolytic assay. Crude extracts of E. coli carrying HlyIII, HlyBA, and HlyB were used to determine hemolytic activity. E. coli carrying vector pUC19 alone was used as a control. Sheep erythrocytes were used in the liquid hemolytic assay as target cells. The release of hemoglobin was measured in the supernatant at A540. Symbols: ▵, HlyIII; •, HlyBA; □, HlyA; ▪, HlyB; ○, HlyA plus HlyB; ▴, pUC19. A lysis assay was carried out by mixing 0.5 ml of crude extract, normalized to 2 mg/ml of protein, with 0.5 ml of the erythrocytes suspension (see Materials and Methods for details). Data are the averages for three experiments. All the standard errors were less than 5% and were therefore not included on the graph for clarity.
Protein-protein interaction between HlyA and HlyB.
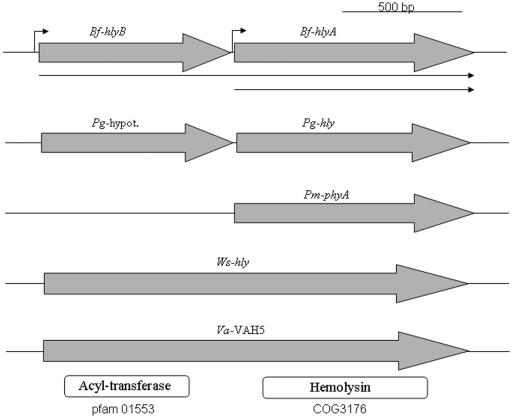
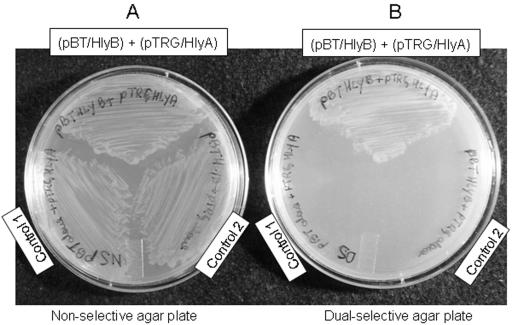
The alignment of HlyA and HlyB amino acid sequences with other bacterial hemolysins revealed that HlyB was homologous (34% identity and 48.5% similarity) to the N-terminal half of Wolinella succinogenes hemolysin and Vibrio anguillarum VAH5 (34), while HlyA was homologous (33% identity and 45% similarity) to the C-terminal region (Fig. 6). HlyA also showed strong homology to Porphyromonas gingivalis hemolysin (58% identity and 69% similarity) and Prevotella melaninogenica hemolysin PhyA (68% identity and 77.5% similarity). In Fig. 6 we show the genetic organization of HlyA and HlyB, which are both similar to V. anguillarum VAH5 and W. succinogenes hemolysins, indicating that they might interact as if they were a large single protein unit. These findings led us to think that HlyA and HlyB would likely have the potential to form protein-protein interactions. Thus, in order to further investigate this hypothesis, experiments using the bacterial two-hybrid system assay were carried out to determine whether HlyA and HlyB would be able to form a complex in vivo. The data shown in Fig. 7 confirm that the HlyB and HlyA form a complex in vivo. Strains containing both the “bait” (pBT/HlyB) and “prey” (pTRG/HlyA) constructs were able to grow on the selective and dual-selective agar plate. In contrast, when the reporter strain was carrying either empty “bait” or empty “prey,” as control for self-activation, there was no growth on the selective medium. This indicates that an interaction between HlyA and HlyB occurred and activated the two-hybrid system, allowing for growth on the selective reporter medium. Taken together, the data presented above and in Fig. 3, 4, and 5 provide evidence that the enhanced hemolytic activity of HlyA and HlyB together is due to the formation of a protein-protein complex.
FIG. 6.
Diagram showing the genetic organization of B. fragilis (Bf) hemolysin genes hlyB and hlyA with other bacterial hemolysin orthologs. Pg-hypot., Porphyromonas gingivalis W83 conserved hypothetical protein (GenBank accession no. AAQ66862); Pg-hly, P. gingivalis W83 hemolysin A (AAQ66861); Pm-phyA, Prevotella melaninogenica hemolysin PhyA (AAB88217); Ws-hly, Wolinella succinogenes DSM 1740 hemolysin (NP_908080); Va-VAH5, Vibrio anguillarum hemolysin VAH5 (AB189398). The gray arrows indicate the open reading frames and their respective direction of transcription. The thin arrows depict mRNA transcript length. The bent arrow depicts the putative promoter region derived from Fig. 9. The conserved motif domains pfam01553 (acyltransferase) and COG3176 (hemolysin) present in respective gene products are depicted as boxes underneath the panel and were obtained from a search in the collection of multiple-sequence alignments at http://www.ncbi.nlm.nih.gov/.
FIG. 7.
E. coli two-hybrid system assay showing protein-protein interaction between HlyB and HlyA. E. coli reporter strain (Stratagene, La Jolla, CA) cotransformed with pBT-HlyB (bait) and pTRG-HlyA (prey) constructs were grown on (A) nonselective plate and (B) dual-selective plate. Self-activation controls are E. coli two-hybrid system reporter strain carrying the following constructs: control 1, empty “bait” (pBT alone) cotransformed with loaded “prey” (pTRG/HlyA); control 2, loaded “bait” (pBT/HlyB) cotransformed with empty “prey” (pTRG alone). See Materials and Methods for details on the bacterial two-hybrid system assay.
Characterization of hlyBA and hlyIII deletion mutants.
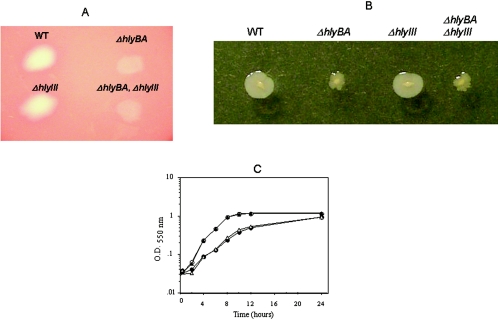
When the hlyBA mutant strain was grown on BAP, hemolytic activity was greatly diminished compared to the parent strain (Fig. 8A). In contrast, the hlyIII mutant strain was not significantly different from the parent strain hemolytic zone. The hlyBA hlyIII double mutant had a diminished, but not abolished, hemolytic zone similar to the hlyBA mutant. These findings show that the HlyBA hemolysin was responsible in large part for the B. fragilis hemolytic activity on BAP. This also suggests that a hemolysin(s) other than HlyBA and HlyIII is still expressed and is yet to be further characterized. In addition, the hlyBA strain had a growth deficiency in SDM medium compared to both the parent and hlyIII strain (Fig. 8C). The growth rate of the hlyBA hlyIII double mutant was nearly identical to that of the hlyBA strain. No significant difference in growth rate was observed when bacteria were grown on rich broth medium (data not shown). These findings correlate with the data presented previously showing that B. fragilis had a strong hemolytic activity on DM-BAP compared to BHIS-BAP. Additional evidence that a disruption in HlyBA activity affects the normal physiological homeostasis of B. fragilis was the effect on colony morphology. When spotted on the surface of a BHIS agar plate, the hlyBA mutant had a significant modification in colony morphology compared to the parent strain (Fig. 8B). hlyBA mutant colonies were irregular and small in contrast to the circular and regular edges observed in the parent strain. Lack of HlyIII did not apparently alter B. fragilis colony morphology.
FIG. 8.
Characterization of hlyBA and hlyIII mutants. A) Hemolytic activity produced by B. fragilis strains. Bacteria were grown on BHIS agar supplemented with 5% human defibrinated blood. Plates were incubated for 8 days anaerobically. Bacteria were removed from the agar surface with sterile cotton swab for clarity. B) Colony morphology of B. fragilis strains grown on agar. Each strain was inoculated on top of BHIS agar and grown under anaerobic conditions. Strain designations are depicted above the colonies in panels A and B (WT, wild-type 638R parent strain). C) Growth of B. fragilis strains on SDM. Bacteria were inoculated into fresh SDM from an overnight culture, and growth rate was measured at an optical density (O.D.) of 550 nm. Data presented are averages from two independent experiments. Symbols: ○, 638R parent strain; •, ΔhlyBA mutant strain; ▴, ΔhlyIII mutant strain; ▵, ΔhlyBA ΔhlyIII double mutant strain.
Effect of iron availability on hlyA and hlyB expression.
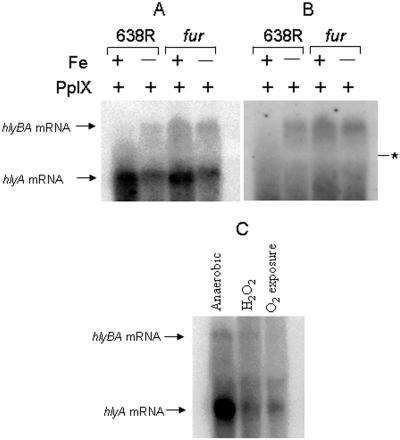
As a first step to understanding regulation of B. fragilis hemolysin expression, hlyBA was used as a model. The hlyBA operon is located immediately downstream from an iron-regulated heme uptake outer membrane protein homolog similar to V. cholerae HutA (16; also data not shown). Therefore, it was plausible that hlyBA regulation was affected by iron availability. To study this, a furA null mutant was constructed and the expression of hlyA and hlyB mRNA was analyzed under iron-replete and iron-limiting conditions. Northern blot hybridization analysis revealed that hlyBA was expressed as a bicistronic mRNA and up-regulated about 2.5-fold under low-iron conditions in the parent strain. In contrast, the normal iron-regulated expression of hlyBA mRNA in the parent strain was no longer repressed under high-iron conditions in the fur mutant (Fig. 9A and B). Interestingly, hlyA was also expressed as a single mRNA in iron-rich medium, but hlyA mRNA expression was repressed approximately threefold under iron-limiting conditions in a Fur-independent manner. This suggests that hlyBA and hlyA mRNAs are differentially regulated by iron and that hlyA alone is also regulated by an unidentified iron-dependent regulator.
FIG. 9.
Autoradiograph of Northern hybridization of total RNA. A and B) B. fragilis 638R and isogenic fur mutant were grown to mid-log phase in SDM containing 100 μM FeSO4 for iron-replete conditions (+Fe) and SDM containing 50 μM 2,2′-bipyridyl and 100 μM desferrioxamine to impose iron-limiting conditions (−Fe). Protoporphyrin IX (PpIX) was added to the medium as a source of tetrapyrrole macrocycle. The probe was an internal fragment from hlyA (panel A) or hlyB (panel B). The positions of the hlyA and hlyB mRNA components are indicated. The asterisk depicts the 16S compression region on the filter membrane. (C) B. fragilis 638R was grown in SDM supplemented with 5 μg/ml hemin and exposed to different oxidative stress conditions. The probe was an internal fragment of hlyA.
Effects of oxidative stress on hlyA and hlyB expression.
To determine whether HlyA and HlyB are regulated by a change in the cellular redox balance, cultures were exposed to atmospheric oxygen and to the oxidant hydrogen peroxide. Figure 9C shows that the hlyA single RNA was expressed under anaerobic conditions, but its level was reduced about threefold following oxygen exposure and treatment with H2O2. The low level of the polycistronic hlyBA transcript was also reduced under the same conditions. Taken together, these results suggest that an oxidative stress insult affects the control of B. fragilis hlyBA and hlyA transcription.
DISCUSSION
In this study we describe the presence of hemolysins in B. fragilis, which is currently classified taxonomically as a nonhemolytic bacterium (18, 19, 53). The presence of at least 10 hemolysin orthologs in the B. fragilis genome suggests that they may play an important role in extraintestinal infections and possibly in the ecology and physiology of the intestinal tract. To the best of our knowledge, there is no previous report on the characterization of hemolysins in B. fragilis despite the fact that rare clinical strains have been reported to have a hemolytic phenotype in vitro (18, 19). Hemolysins/cytolysins are important toxins that lyse neutrophils and other host cells which can affect the immune response of the host and also provide a source of nutrients for the invading microorganism (13, 24, 38, 51). Hemolysins/cytolysins in many bacteria are not only considered potent virulence factors but are also cytolytic/bactericidal factors used to inhibit other prokaryotes and eukaryotes in highly competitive ecological systems (9). A classical example of the participation of cytolysin in both pathogenesis and ecological competitiveness is the two-peptide component cytolysin (bacteriocin) of the gastrointestinal tract inhabitant Enterococcus faecalis. Production of cytolysin by E. faecalis enhances lethality in human infections and animal models as well as providing a competitive advantage over other gram-positive bacteria in the intestinal tract (8, 9). In this regard, it is possible that the extensive number of hemolysins/cytolysins in Bacteroides species not only play a role in the infectious process but they also might contribute to their ability to dominate and take over the colonization of the human large intestinal tract.
The findings presented in this study indicate that hemolysins HlyA and HlyB are a two-component cytolysin. This is based on results showing that when both components are expressed together, there is a synergistic effect on hemolytic activity, and there is a protein-protein interaction. However, it is not known whether HlyA and HlyB protein complex formation occurs at the cell membrane or occurs prior to attachment on the eukaryotic cell surface. Though two-component hemolysins are produced by several bacteria (9, 25, 50, 52), B. fragilis HlyBA does not share homology with the classical Serratia marcescens family of two-component hemolysins (17) or bacterial pore-forming toxins of the RTX (repeat-in-toxin) exoprotein family (46). The mechanism of hemolytic activity of HlyBA is not known, but because HlyB has a conserved phospholipid/glycerol acyltransferase superfamily motif, we think that it might possess functional features within this group of enzymes involved in phospholipid biosynthesis (28, 48). Interestingly, the HlyB and HlyA peptides are homologous, respectively, to the acyltransferase and the hemolytic domains of hemolysins found in microorganisms such as Vibrio anguillarum and W. succinogenes. It is possible that a genetic mutation might have occurred in Bacteroides and in the closely related anaerobe P. gingivalis where HlyA and HlyB resulted from a split of a single peptide unit into two distinct peptides. The reverse phenomenon cannot be ruled out either. The reason for this is not known, but we presume that the two separate peptides may have other additional physiological properties directly interfering with bacterial physiology. In support of this is the fact that the B. thetaiotaomicron hlyBA genes are arranged in a transcriptional fashion similar to that in B. fragilis and the levels of B. thetaiotaomicron hlyBA mRNA are more abundant in early growth phase than the levels in later growth phase in vitro (44; NCBI/Gene Expression Omnibus (GEO) database repository/GEO database entry accession no. GSE2231).
Expression of HlyB and HlyA was shown to be differentially and coordinately regulated by oxygen and iron availability. The former represses hlyBA mRNA upon exposure to aerobic conditions, and our hypothesis is that HlyBA may be a potential virulence factor used by B. fragilis to injure and lyse host tissue cells in order to obtain essential nutrients in an extraintestinal anaerobic environment, such as an abscess. This is consistent with the fact that B. fragilis is able to proliferate after the establishment of anaerobic conditions at the site of infection (35, 36, 37, 39). In general, anaerobic conditions in the infected tissues are formed by the consumption of oxygen by facultative bacteria often found associated with anaerobes in polymicrobial infections (35, 36, 37, 39). The effects of oxygen limitation on hemolysin production and other virulence factors have been demonstrated in several pathogenic bacteria (10, 27, 47). For example, E. faecalis cytolysin/hemolysin CylLL and CylLS are up-regulated (8.6-fold) at the transcriptional level under anaerobic conditions compared to the levels in the presence of oxygen (10). The implications of anaerobic regulation of HlyBA in the pathogenicity of B. fragilis correlate nicely with the fact that the establishment of anaerobic infections in extraintestinal tissues follows the depletion of oxygen at the site of infection. Therefore, it is likely that HlyBA may contribute to B. fragilis pathogenicity in a low-oxygen and low-iron environment as discussed below.
Several studies have shown that B. fragilis is unable to synthesize protoporphyrin macrocycle and has an essential requirement for heme and nonheme iron (31, 45). This nutritional requirement correlates with the expression and role HlyBA might play to make possible bacterial access to iron and heme under the low-iron conditions encountered in host tissues. The production of hemolysins/cytolysins has been associated with the ability of pathogenic bacteria to obtain heme as a source of iron from lysed erythrocytes and other host cells (13). In agreement, iron-limiting conditions regulate the synthesis of hemolysins in many pathogenic bacteria (2, 3, 6, 23, 29). Thus, the iron-dependent regulation of hlyBA mRNA may help B. fragilis to overcome the iron-limiting conditions imposed by a host's iron-withholding mechanisms.
In conclusion, we showed in this study that B. fragilis encodes several functional hemolysins not previously characterized in this organism. Our studies on the synthesis and regulation of hemolysins may help us to understand B. fragilis pathogenicity and why this organism has a much greater potential to cause infections than any other anaerobic species that colonizes the human intestinal tract. In support of the hypothesis that hemolysins are important for B. fragilis virulence is a recent comparative genome analysis suggesting that the presence of hemolysin-like proteins and capsular polysaccharides may explain the difference in pathogenic potentials between the two closely related species B. fragilis and B. thetaiotaomicron (22).
Acknowledgments
We thank the Sanger Institute for the release of sequence data produced by the B. fragilis Sequencing Group at the Sanger Institute, which can be obtained from ftp//ftp.sanger.ac.uk/pub/pathogens/bf/. We also thank B. E. Uhlin (Umea University, Sweden) for kindly providing E. coli YMZ19 (clyA::kan) strain and E. C. Pesci (East Carolina University) for critical review of the manuscript.
This work was supported in part from PHS grant AI40588 to C.J.S.
Editor: F. C. Fang
REFERENCES
- 1.Bernheimer, A. W. 1988. Assay of hemolytic toxins. Methods Enzymol. 165:213-217. [DOI] [PubMed] [Google Scholar]
- 2.Braun, V. 1985. Iron supply as a virulence factor, p. 168-176. In G. G. Jackson and H. Thomas (ed.), The pathogenesis of bacterial infections. Bayer Symposium III. Springer-Verlag, Berlin, Germany.
- 3.Braun, V., and T. Focareta. 1991. Pore-forming bacterial protein hemolysins (cytolysins). Crit. Rev. Microbiol. 18:115-158. [DOI] [PubMed] [Google Scholar]
- 4.Brook, I. 1989. Pathogenicity of Bacteroides fragilis group. Ann. Clin. Lab. Sci. 19:360-376. [PubMed] [Google Scholar]
- 5.Brook, I., and E. H. Frazier. 2000. Aerobic and anaerobic microbiology in intra-abdominal infections associated with diverticulitis. J. Med. Microbiol. 49:827-830. [DOI] [PubMed] [Google Scholar]
- 6.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerdeno-Tarraga, A. M., S. Patrick, L. C. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. A. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. T. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463-1465. [DOI] [PubMed] [Google Scholar]
- 8.Coburn, P. S., and M. S. Gilmore. 2003. The Enterococcus faecalis cytolysin: a novel toxin active against eukaryotic and prokaryotic cells. Cell. Microbiol. 5:661-669. [DOI] [PubMed] [Google Scholar]
- 9.Cox, C. R., P. S. Coburn, and M. S. Gilmore. 2005. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr. Protein Pept. Sci. 6:77-84. [DOI] [PubMed] [Google Scholar]
- 10.Day, A. M., J. H. Cove, and M. K. Phillips-Jones. 2003. Cytolysin gene expression in Enterococcus faecalis is regulated in response to aerobiosis conditions. Mol. Genet. Genomics 269:31-39. [DOI] [PubMed] [Google Scholar]
- 11.Finegold, S. M., and W. L. George. 1989. Anaerobic infections in humans. Academic Press, San Diego, CA.
- 12.Foulon, I., D. Pierard, G. Muyldermans, K. Vandoorslaer, O. Soetens, P. Rosseel, and S. Lauwers. 2003. Prevalence of fragilysin gene in Bacteroides fragilis isolates from blood and other extraintestinal samples. J. Clin. Microbiol. 41:4428-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, F. C., III, A. O. Tzianabos, and A. D. Onderdonk. 1996. The capsular polysaccharide complex of Bacteroides fragilis induces cytokine production from human and murine phagocytic cells. Infect. Immun. 64:1065-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson, F. C., III, A. B. Onderdonk, D. L. Kasper, and A. O. Tzianabos. 1998. Cellular mechanism of intraabdominal abscess formation by Bacteroides fragilis. J. Immunol. 160:5000-5006. [PubMed] [Google Scholar]
- 16.Henderson, D. P., and S. M. Payne. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 176:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertle, R. 2005. The family of Serratia type pore forming toxins. Curr. Protein Pept. Sci. 6:313-325. [DOI] [PubMed] [Google Scholar]
- 18.Holdeman, L. M., E. P. Cato, and W. E. C. Moore. 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 19.Holdeman, L. V., R. W. Kelley, and W. E. C. Moore. 1984. Family I. Bacteroidaceae Pribam 1933, 10AL, p. 602-631. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 20.Hooper, L. V., T. Midtvedt, and J. L. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 21.Krinos, C. M., M. J. Coyne, K. G. Weinacht, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555-558. [DOI] [PubMed] [Google Scholar]
- 22.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menestrina, G., M. Dalla Serra, M. Comai, M. Coraiola, G. Viero, S. Werner, D. A. Colin, H. Monteil, and G. Prevost. 2003. Ion channels and bacterial infection: the case of beta-barrel pore-forming protein toxins of Staphylococcus aureus. FEBS Lett. 552:54-60. [DOI] [PubMed] [Google Scholar]
- 25.Miles, G., S. Cheley, O. Braha, and H. Bayley. 2001. The staphylococcal leukocidin bicomponent toxin forms large ionic channels. Biochemistry 40:8514-8522. [DOI] [PubMed] [Google Scholar]
- 26.Moore, W. E., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouriño, M., F. Muñoa, C. Balsalobre, P. Diaz, C. Madrid, and A. Juarez. 1994. Environmental regulation of α-haemolysin expression in Escherichia coli. Microb. Pathog. 16:249-259. [DOI] [PubMed] [Google Scholar]
- 28.Neuwald, A. F. 1997. Barth syndrome may be due to an acyltransferase deficiency. Curr. Biol. 7:R465-R466. [DOI] [PubMed] [Google Scholar]
- 29.Poole, K., and V. Braun. 1988. Iron regulation of Serratia marcescens hemolysin gene expression. Infect. Immun. 56:2967-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Privitera, G., A. Dublanchet, and M. Sebald. 1979. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J. Infect. Dis. 139:97-101. [DOI] [PubMed] [Google Scholar]
- 31.Rocha, E. R., M. de Uzeda, and J. H. Brock. 1991. Effect of ferric and ferrous iron chelators on growth of Bacteroides fragilis under anaerobic conditions. FEMS Microbiol. Lett. 68:45-50. [DOI] [PubMed] [Google Scholar]
- 32.Rocha, E. R., and C. J. Smith. 1997. Regulation of Bacteroides fragilis katB mRNA expression by oxidative stress and carbon limitation. J. Bacteriol. 179:7033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rocha, E. R., and C. J. Smith. 2004. Transcriptional regulation of the Bacteroides fragilis ferritin gene (ftnA) by redox stress. Microbiology 150:2125-2134. [DOI] [PubMed] [Google Scholar]
- 34.Rodkhum, C., I. Hirono, J. H. Crosa, and T. Aoki. 2005. Four novel hemolysin genes of Vibrio anguillarum and their virulence to rainbow trout. Microb. Pathog. 39:109-119. [DOI] [PubMed] [Google Scholar]
- 35.Rotstein, O. D. 1993. Interactions between leukocytes and anaerobic bacteria in polymicrobial surgical infections. Clin. Infect. Dis. 16(Suppl. 4):S190-S194. [DOI] [PubMed] [Google Scholar]
- 36.Rotstein, O. D., T. L. Pruett, and R. L. Simmons. 1985. Mechanisms of microbial synergy in polymicrobial surgical infections. Rev. Infect. Dis. 7:151-170. [DOI] [PubMed] [Google Scholar]
- 37.Rotstein, O. D., and J. Kao. 1988. The spectrum of Escherichia coli-Bacteroides fragilis pathogenic synergy in an intraabdominal infection model. Can. J. Microbiol. 34:352-357. [DOI] [PubMed] [Google Scholar]
- 38.Rowe, G. E., and R. A. Welch. 1994. Assays of hemolytic toxins. Methods Enzymol. 235:657-667. [DOI] [PubMed] [Google Scholar]
- 39.Sawyer, R. G., M. D. Spengler, R. B. Adams, and T. L. Pruett. 1991. The peritoneal environment during infection. The effect of monomicrobial and polymicrobial bacteria on pO2 and pH. Ann. Surg. 213:253-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sears, C. L. 2001. The toxins of Bacteroides fragilis. Toxicon 39:1737-1746. [DOI] [PubMed] [Google Scholar]
- 41.Shoemaker, N. B., C. Getty, J. F. Gardner, and A. A. Salyers. 1986. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J. Bacteriol. 165:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, C. J., L. A. Rollins, and A. C. Parker. 1995. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid 34:211-222. [DOI] [PubMed] [Google Scholar]
- 43.Smith, C. J., E. R. Rocha, and B. J. Paster. 2003. The medically important Bacteroides spp. in health and disease. In M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, N.Y. [Online.] http://link.springer-ny.com/link/service/books/10125/ny.
- 44.Sonnenburg, J. L., J. Xu, D. D. Leip, C. H. Chen, B. P. Westover, J. Weatherford, J. D. Buhler, and J. I. Gordon. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955-1959. [DOI] [PubMed] [Google Scholar]
- 45.Sperry, J. F., M. D. Appleman, and T. D. Wilkins. 1977. Requirement of heme for growth of Bacteroides fragilis. Appl. Environ. Microbiol. 34:386-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanley, P., V. Koronakis, and C. Hughes. 1998. Acylation of Escherichia coli hemolysin: a unique protein lipidation mechanism underlying toxin function. Microbiol. Mol. Biol. Rev. 62:309-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiwari, R. P., K. Deol, P. Rishi, and J. S. Grewal. 2002. Factors affecting haemolysin production and congo red binding in Salmonella enterica serovar Typhimurium DT 98. J. Med. Microbiol. 51:503-509. [DOI] [PubMed] [Google Scholar]
- 48.Turnbull, A. P., J. B. Rafferty, S. E. Sedelnikova, A. R. Slabas, T. P. Schierer, J. T. M. Kroon, J. W. Simon, T. Fawcett, I. Nishida, N. Murata, and D. W. Rice. 2001. Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase. Structure 9:347-353. [DOI] [PubMed] [Google Scholar]
- 49.Wai, S. N., M. Westermark, J. Oscarsson, J. Jass, E. Maier, R. Benz, and B. E. Uhlin. 2003. Characterization of dominantly negative mutant ClyA cytotoxin proteins in Escherichia coli. J. Bacteriol. 185:5491-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weikel, C. S., F. D. Grieco, J. Reuben, L. L. Myers, and R. B. Sack. 1992. Human colonic epithelial cells, HT29/C1, treated with crude Bacteroides fragilis enterotoxin dramatically alter their morphology. Infect. Immun. 60:321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welch, R. A. 1991. Pore-forming cytolysins of gram-negative bacteria. Mol. Microbiol. 5:521-528. [DOI] [PubMed] [Google Scholar]
- 52.Williams, M. L., and M. L. Lawrence. 2005. Identification and characterization of a two-component hemolysin from Edwardsiella ictaluri. Vet. Microbiol. 108:281-289. [DOI] [PubMed] [Google Scholar]
- 53.Wren, M. W. D. 1991. Laboratory diagnosis of anaerobic infection, p. 180-196. In B. I. Duerden and B. S. Drasar (ed.), Anaerobes in human disease. Wiley-Liss, New York, N.Y.
- 54.Xu, J., and J. L. Gordon. 2003. Honor thy symbionts. Proc. Natl. Acad. Sci. USA 100:10452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]