Abstract
Objectives: To detect the serum proteomic patterns by using SELDI-TOF-MS (surface enhanced laser desorption/ionization-time of flight-mass spectrometry) technology and CM10 ProteinChip in colorectal cancer (CRC) patients, and to evaluate the significance of the proteomic patterns in the tumour staging of colorectal cancer. Methods: SELDI-TOF-MS and CM10 ProteinChip were used to detect the serum proteomic patterns of 76 patients with colorectal cancer, among them, 10 Stage I, 19 Stage II, 16 Stage III and 31 Stage IV samples. Different stage models were developed and validated by support vector machines, discriminant analysis and time-sequence analysis. Results: The Model I formed by 6 protein peaks (m/z: 2759.58, 2964.66, 2048.01, 4795.90, 4139.77 and 37761.60) could be used to distinguish local CRC patients (Stage I and Stage II) from regional CRC patients (Stage III) with an accuracy of 86.67% (39/45). The Model II formed by 3 protein peaks (m/z: 6885.30, 2058.32 and 8567.75) could be used to distinguish locoregional CRC patients (Stage I, Stage II and Stage III) from systematic CRC patients (Stage IV) with an accuracy of 75.00% (57/76). The Model III could distinguish Stage I from Stage II with an accuracy of 86.21% (25/29). The Model IV could distinguish Stage I from Stage III with accuracy of 84.62% (22/26). The Model V could distinguish Stage II from Stage III with accuracy of 85.71% (30/35). The Model VI could distinguish Stage II from Stage IV with accuracy of 80.00% (40/50). The Model VII could distinguish Stage III from Stage IV with accuracy of 78.72% (37/47). Different stage groups could be distinguished by the two-dimensional scattered spots figure obviously. Conclusion: This method showed great success in preoperatively determining the colorectal cancer stage of patients.
Keywords: Colorectal cancer, SELDI-TOF-MS (surface enhanced laser desorption/ionization-time of flight-mass spectrometry), Staging, Bio-informatics
INTRODUCTION
Worldwide, colorectal cancer is the third most frequently occurring cancer in both sexes; it ranks second in developed countries (Hawk and Levin, 2005). In China, colorectal cancer is the fourth leading cause of cancer mortality in big cities, the fifth in the countryside, has been a major cancer and will be increasing in the near future (Zheng and Cai, 2003). The lack of good serum tumor markers for colorectal cancer makes it very difficult to determine its preoperative molecular stage. By far, determining of colorectal cancer stage can only be possible after complete surgical resection rather than after a presurgical biopsy. It is very important to oncologists to comprehend the tomor margin before surgery in order to choose rational surgical schemes and formulate rational treatment plans. It has been confirmed that the neoadjuvent and adjuvant therapy are only beneficial to some selected colorectal cancer patients.
In proteomics, the technology of micro-array and bio-informatics carved out a new way to effectively seek tumor markers. Surface enhanced laser desorption/ionization-time of flight-mass spectrometry (SELDI-TOF-MS) is such a new path to provide high-throughput protein profiling (Merchant and Weinberger, 2000). In this study the proteomics patterns in sera of different stages of colorectal cancer patients were detected by SELDI-TOF-MS technology. The serum protein patterns of colorectal cancer patients before surgery were obtained to build the stage models whose protein serum pattern was different in different colorectal cancer stages. This method could be helpful for discrimination of different stages before operation. And we also differentiated patients with liver metastasis of Stage IV from those of local celiac metastasis and distant metastasis.
MATERIALS AND METHODS
Sample preparation
All the samples were obtained from patients in the Second Affiliated Hospital, School of Medicine, Zhejiang University, after informed consent. The sera of the patients were obtained before any therapeutic measures were implemented. The diagnoses were confirmed by postsurgical pathology. The total colorectal cancer group consisted of 76 cases at different clinical stages (the International Union Against Cancer, UICC, 1997), Stage I (n=10), Stage II (n=19), Stage III (n=16) and Stage IV (n=31). The median age of the colorectal cancer patients was 57 years (range, 18~89 years). There were 54 males and 22 females. The median ages of the Stages I, II, III and IV groups were 53 years (range, 37~77 years), 64 years (range, 32~77 years), 58 years (range, 36~71 years) and 55 years (range, 18~89 years) respectively. All the samples were collected in the early morning before breakfast, and then the sera were separated at once and stored at −80 °C until use.
Analysis of ProteinChip array
After thawing and 2 min of centrifugation (10 000 r/min), 5 μl serum sample was added into 10 μl 0.5% U9 (9 mol/L urea, 0.2% CHAPS (3[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate), 0.1% DTT (DL-dithiothreitol)) in a 96-well plate and incubated for 30 min at 4 °C with 600 r/min vigorous shaking. The ProteinChip array cassette was put into a 96-well bioprocessor and 200 μl NaAc (50 mmol/L, pH 4.0) was put into each well, and incubated for 5 min at 4 °C with 600 r/min vigorous shaking. The liquid was removed. The procedure was repeated once. Then 185 μl NaAc was added into each well in the 96-well plate (600 r/min, 2 min) and then 100 μl samples disposed above of different patients were added into different well separately of the ProteinChip array cassette (600 r/min, 1 h). After the content from each well was removed, each well was washed with 200 μl NaAc (pH=4.0, 600 r/min, 5 min). The procedure was repeated two more times. Each spot was washed with 200 μl HPLC water, which was removed immediately. The procedure was repeated once. After air drying, 1 μl SPA (sinapic acid) was applied to each spot. After air drying for 5 min, another 1 μl SPA was applied. Allow air-drying.
Ciphergen SELDI Protein Biology System II plus (PBS II plus) and ProteinChip Software (Version 3.2, Ciphergen Biosystems) were used to read the chips and analyze the data. The following settings were used: laser intensity 165, 65 laser shots per sample, detector sensitivity 7, automatically detected peaks from 2 000 to 30 000 m/z (mass to charge ratios). Mass accuracy was calibrated to less than 0.1% using the All-in-1 peptide molecular mass standard (Ciphergen Biosystems). The peaks were normalized and noises were filtrated (first signal to noise ratio>2.5). Peak clusters were completed using second-pass peak selection (signal to noise ratio>2, within 0.3% mass window) and estimated peaks were added. The biomarker wizard of ProteinChip Software 3.2 was used to compare the data of different group by Mann-Whitney U test and discrepant mass peaks were found.
Statistical analysis
SELDI-TOF-MS technology and CM10 ProteinChip (weak cation exchange) were used to detect the serum proteomic patterns of colorectal cancer. The samples of different stage groups models were developed and validated by support vector machines, discriminant analysis and time-sequence analysis. These statistical analysis tools were implemented by MATLAB-NNTools software. Training was conducted to convergence on the training data and minimized the errors.
Discriminant analysis and support vector machines models introduced random perturbations in multiple runs to test the consistency of the top 10 ranked peaks, measured by the P value of m/z peaks of computed ranks from multiple runs. Then stage models were built using the selected peaks. The models established based on these selected biomarkers should be further validated independently. In such studies, validation datasets preferably should be from sources different from that of the original training dataset. This is one way to ensure that the performance of the selected biomarkers is not influenced by systematic biases between different groups. Time-sequence analysis was used to distinguish different stage groups.
Here, leave-one-out cross-validation approach was applied to estimate the accuracy of the classifier to determine the misclassification rate. For each step of the cross-validation, one sample was left out. The possibility of obtaining a small cross-validated misclassification rate by chance was obtained by repeating the entire cross-validation procedure using n random permutations of the class labels for the clinical criteria being evaluated.
RESULTS
1. Comparison of local (Stages I and II, 29 cases) CRC with regional CRC (Stage III, 16 cases)
Totally, 11 qualified and discrepant mass peaks were detected in the comparison of local CRC with regional CRC. Through leave-one-out cross-validation, discriminant analysis screening out 6 protein mass peaks (m/z: 2759.58, 2964.66, 2048.01, 4795.90, 4139.77 and 37761.60) to build the Model I, which could develop and evaluate the peptide patterns for distinguishing local CRC patients from regional CRC patients. The accuracy of the blind prediction was 86.67% (39/45). The sensitivity and specificity of Model I were 86.21% (25/29) and 87.50% (14/16) respectively (Table 1 and Table 2).
Table 1.
The 6 selected discrepant protein mass peaks comparison of local CRC with regional CRC of Model I
| Protein peaks (m/z) | Local CRC (mean±SD) | Regional CRC (mean±SD) | P value (t-test) |
| 2759.58 | 1.189±0.622 | 1.946±0.832 | 0.003 |
| 2964.66 | 1.650±0.629 | 1.048±0.809 | 0.006 |
| 2048.01 | 1.073±0.862 | 1.926±1.290 | 0.128 |
| 4795.90 | 1.745±0.756 | 2.440±0.837 | 0.021 |
| 4139.77 | 1.900±0.602 | 2.381±0.732 | 0.023 |
| 37761.60 | 0.020±0.016 | 0.030±0.012 | 0.031 |
Table 2.
The cross-validation blind test results of the test set in Model I (cases)
| Groups | Local CRC | Regional CRC | Total |
| Local CRC (predicted) | 25 (86.21%) | 2 (13.79%) | 27 |
| Regional CRC (predicted) | 4 (12.50%) | 14 (87.50%) | 18 |
| Total | 29 | 16 | 45 |
Sensitivity: 86.21% (25/29); Specificity: 87.50% (14/16); Accuracy: 86.67% (39/45)
2. Comparison of locoregional (Stages I, II and III, 45 cases) CRC with systemic CRC (Stage IV, 31 cases)
Totally, 24 qualified and discrepant mass peaks were detected in the comparison of locoregional CRC with systemic CRC. Through leave-one-out crossvalidation, discriminant analysis screened out 3 protein mass peaks (m/z: 6885.30, 2058.32 and 8567.75) to build the Model II, which could develop and evaluate the peptide patterns for distinguishing locoregional CRC patients from systemic CRC patients. The accuracy of the blind prediction was 75.00% (57/76). The sensitivity and specificity of Model II were 71.11% (32/45) and 80.65% (25/31) respectively (Table 3 and Table 4).
Table 3.
The 3 selected discrepant protein mass peaks comparison of locoregional CRC with systemic CRC of Model II
| Protein peaks (m/z) | Locoregional CRC (mean±SD) | Systemic CRC (mean±SD) | P value (t-test) |
| 6885.30 | 2.382±1.186 | 1.593±1.048 | 0.002 |
| 2058.32 | 0.585±0.852 | 1.178±0.831 | 0.003 |
| 8567.75 | 6.865±3.660 | 4.668±2.400 | 0.006 |
Table 4.
The cross-validation blind test results of the test set in Model II (cases)
| Groups | Locoregional CRC | Systemic CRC | Total |
| Locoregional CRC (predicted) | 32 (71.11%) | 6 (19.35%) | 38 |
| Systemic CRC (predicted) | 13 (28.89%) | 25 (80.65%) | 38 |
| Total | 45 | 31 | 76 |
Sensitivity: 71.11% (32/45); Specificity: 80.65% (25/31); Accuracy: 75.00% (57/76)
3. According to different protein peaks we built different stage models: III, IV, V, VI, VII (Table 5). The Model III could distinguish Stage I from II with an accuracy of 86.21% (25/29). The Model IV could distinguish Stage I from III with an accuracy of 84.62% (22/26). The Model V could distinguish Stage II from III with an accuracy of 85.71% (30/35). The Model VI could distinguish Stage II vs IV with an accuracy of 80.00% (40/50). The Model VII could distinguish Stage III from IV with accuracy of 78.72% (37/47).
Table 5.
The accuracy, specificity, sensitivity and selected discrepant protein mass peaks of Models III to VII
| Model | Groups | m/z | A (%) | Sp (%) | Se (%) |
| III | Stage I vs II | 3376.29; 4285.21; 13762.60 | 86.21 | 100.00 | 78.95 |
| IV | Stage I vs III | 2759.58; 2964.66 | 84.62 | 80.00 | 87.50 |
| V | Stage II vs III | 4139.77; 2292.31; 2759.58 | 85.71 | 84.21 | 87.50 |
| VI | Stage II vs IV | 2954.65; 4795.90; 2174.44; 2890.47 | 80.00 | 78.95 | 80.65 |
| VII | Stage III vs IV | 6634.67; 2022.55 | 78.72 | 81.25 | 77.42 |
m/z: Mass/charge; A: Accuracy; Sp: Specificity; Se: Sensitivity
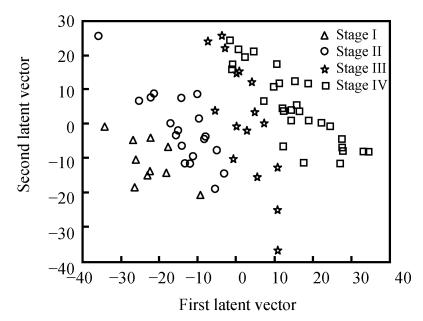
4. The two-dimensional scattered spots figure of different stage groups (Fig.1). Different stage groups could be obviously distinguished by the two-dimensional scattered spots figure.
Fig. 1.
The two-dimensional scattered spots figure of different stage groups
DISCUSSION
Many patients had evidence of locally advanced or metastatic colorectal cancer at the time of initial presentation; only half of those who underwent apparently curative resection survived 5 years (Harris et al., 2002; Ries et al., 2000; Greenlee et al., 2000). The causative reason of death is associated directly with stage and therapeutic methods. The lack of good serum tumor markers causes great difficulty in determining its molecular stage before colorectal cancer surgery. Several common medical examination tools such as endoscopy, MRI, CT, etc. also cannot differentiate stage accurately. It is very important to oncologists to comprehend the stage before surgery in order to choose surgical schemes and formulate rational plans.
As a new proteomics approach, the ProteinChip based on SELDI-TOF-MS could bind the proteins in the samples unselectively. It combines ProteinChip array with time-of-flight mass spectrometry and offers the advantages of speed, simplicity, sensitivity and suitability for a comparative study. When laser in SELDI fired the bond proteins, they were charged and became ions in the gaseous phase. The ions’ time of flight in the electric field depended on their m/z so the detector could receive them and transform their m/z and amount into peaks of different position and height. It was a proteomics technology of high sensitivity and high throughput (Issaq et al., 2002; Weinberger et al., 2000). In recent years, ProteinChip was successfully utilized in diagnosis of cancer, screening of tumor markers and in other proteomics fields (Paradis et al., 2005; Alexe et al., 2004; Semmes et al., 2005; Chen et al., 2004). Because of the multifactorial nature of cancer, it is very likely that a combination of several markers will be necessary. To look for such “fingerprints” of cancer, it will require not only high-throughput genomic or proteomic profiling, but also sophisticated bioinformatics tools for complex data analysis and pattern recognition (Zhukov et al., 2003). Tons of data could be obtained in one experiment, so the bio-informatics methods such as discriminant analysis, support vector machines (SVM), ANN (artificial neural networks) etc. were needed to collect and analysis them.
This method produced excellent cross-validation accuracy estimates. The leave-one-out method produced the highest accuracy estimate for this dataset.
In this study, the stage proteomics pattern of colorectal cancer was found and VII differentiated models were built by SELDI-TOF-MS ProteinChip and bio-informatics tools. The blind or cross tests of these models showed good accuracies, sensitivities, and specificities in different stage of colorectal cancer. The six newly found biomarkers in Model I might become the potential tumor markers distinguishing local CRC from reginal CRC. Model I can direct neoadjuvant chemotherapy for colorectal cancer. Some reports on combined modality treatment with chemotherapy and radiotherapy before operation clearly showed a downstaging effect on the primary tumor and decreased prevalence of regional involved lymph node (Kotake and Koyama, 1995). In 2005, the Clinical Practice Guidelines in Oncology of American NCCN (national comprehensive cancer network, http://www.nccn.org/) recommended neoadjuvant therapy for regional rectal cancer.
Three protein mass peaks built the Model II which can develop and evaluate the peptide patterns for distinguishing locoregional CRC patients from systemic CRC patients and can distinguish whether CRC patients had distant metastases or not to direct therapy. The Model III can distinguish Stage I from II. The Model IV can distinguish Stage I from III so that it can eliminate lymph nodes micro-metastasis of Stage I. The Model V and Model VI can eliminate lymph nodes or distant metastases of Stage II. The Model VII can eliminate distant metastases of Stage III. The models above can meticulously distinguish different stages of CRC patients before surgery. It is very important for oncologists to master surgical approach, improve the prognosis and carry through comprehensive therapy.
Colorectal cancer staging is currently based solely on simple clinicopathologic features such as bowel wall penetration and lymph node metastasis. Unfortunately, clinical staging systems often fail to distinguish the biologic behavior of a large number of tumors, resulting in the systemic overtreatment or undertreatment of patients with adjuvant therapies. The intermediate stages of II and III are not extremely useful in distinguishing good from poor prognosis patients. It is disputable if Stage II patients should receive adjuvant therapies after surgery. Fig.1 can direct the adjuvant therapies of Stage II patients. Those who were close to Stage III in the two-dimensional scattered spots figure may have high risk factors and should receive adjuvant therapies. Stage III patients who were close to Stage IV predicted poor prognosis and should be followed up closely and treated positively.
Here, the results were analyzed by discriminant analysis, but other bio-informatics methods such as SVM were also tried. Similar sensitivities and specificities could be obtained that proved the stability and reliability of the results.
In conclusion, the SELDI-TOF-MS ProteinChip technology combined with sophisticated bio-informatics tools can lead to the discovery of new biomarkers and establish patterns with high sensitivity and specificity for differentiation of different stages in CRC cancer.
Footnotes
Project (No. 30471987) supported by the National Natural Science Foundation of China
References
- 1.Alexe G, Alexe S, Liotta LA, Petricoin E, Reiss M, Hammer PL. Ovarian cancer detection by logical analysis of proteomic data. Proteomics. 2004;4(3):766–783. doi: 10.1002/pmic.200300574. [DOI] [PubMed] [Google Scholar]
- 2.Chen YD, Zheng S, Yu JK, Hu X. Development of serum biomarker model and its application in colorectal cancer diagnosis. J Tumor Marker Oncol. 2004;19(3):251–258. [Google Scholar]
- 3.Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics 2000. CA Cancer J Clin. 2000;50(1):7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- 4.Harris GJ, Church JM, Senagore AJ, Lavery IC, Hull TL, Strong SA, Fazio VW. Factors affecting local recurrence of colonic adenocarcinoma. Dis Colon Rectum. 2002;45(8):1029–1034. doi: 10.1007/s10350-004-6355-1. [DOI] [PubMed] [Google Scholar]
- 5.Hawk ET, Levin B. Colorectal cancer prevention. J Clin Oncol. 2005;23(2):378–391. doi: 10.1200/JCO.2005.08.097. [DOI] [PubMed] [Google Scholar]
- 6.Issaq HJ, Veenstra TD, Conrads TP, Felschow D. The SELDI-TOF MS approach to proteomics: protein profiling and biomarker identification. Biochem Biophys Res Commun. 2002;292(3):587–592. doi: 10.1006/bbrc.2002.6678. [DOI] [PubMed] [Google Scholar]
- 7.Kotake K, Koyama Y. Neoadjuvant therapy for colorectal cancer. Gan to Kagaku Ryoho. 1995;22(13):1886–1892. [PubMed] [Google Scholar]
- 8.Merchant M, Weinberger SR. Recent advancements in surface-enhanced laser desorption/ionization-time of flight-mass spectrometry. Electrophoresis. 2000;21(6):1164–1177. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1164::AID-ELPS1164>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 9.Paradis V, Degos F, Dargere D, Pham N, Belghiti J, Degott C, Janeau JL, Bezeaud A, Delforge D, Cubizolles M, et al. Identification of a new marker of hepatocellular carcinoma by serum protein profiling of patients with chronic liver diseases. Hepatology. 2005;41(1):40–47. doi: 10.1002/hep.20505. [DOI] [PubMed] [Google Scholar]
- 10.Ries LA, Wingo PA, Miller DS, Howe HL, Weir HK, Rosenberg HM, Vernon SW, Cronin K, Edwards BK. The annual report to the nation on the status of cancer, 1973~1997, with a special section on colorectal cancer. Cancer. 2000;88(10):2398–2424. doi: 10.1002/(SICI)1097-0142(20000515)88:10<2398::AID-CNCR26>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 11.Semmes OJ, Feng Z, Adam BL, Banez LL, Bigbee WL, Campos D, Cazares LH, Chan DW, Grizzle WE, Izbicka E, et al. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem. 2005;51(1):102–112. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- 12.Weinberger SR, Morris TS, Pawlak M. Recent trends in protein biochip technology. Pharmacogenomics. 2000;1(4):395–416. doi: 10.1517/14622416.1.4.395. [DOI] [PubMed] [Google Scholar]
- 13.Zheng S, Cai SR. Colorectal cancer epidemiology and prevention study in China. Chinese-German J Clin Oncol. 2003;2(2):72–75. [Google Scholar]
- 14.Zhukov TA, Johanson RA, Cantor AB, Clark RA, Tockman MS. Discovery of distinct protein profiles specific for lung tumors and pre-malignant lung lesions by SELDI mass spectrometry. Lung Cancer. 2003;40(3):267–279. doi: 10.1016/s0169-5002(03)00082-5. [DOI] [PubMed] [Google Scholar]