Figure 1.
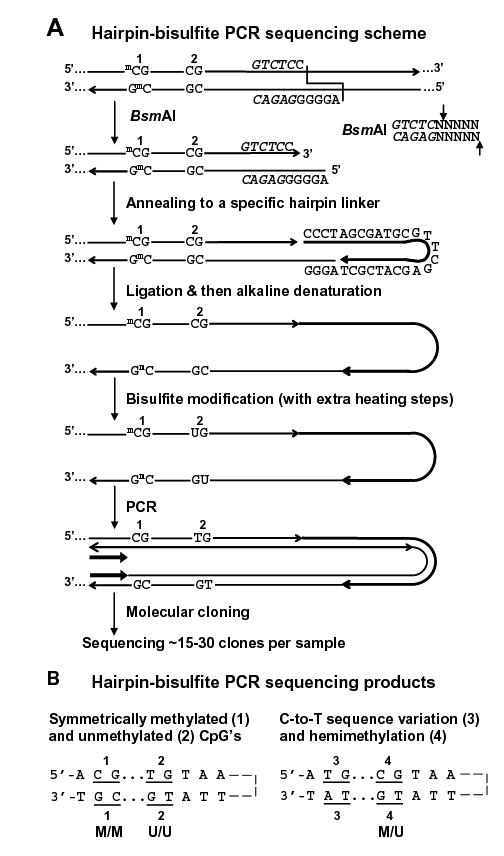
Hairpin genomic sequencing methodology. (A) Hairpin GS of the NBL2 repeat with the covalently linked upper and lower strands (not to scale) diagrammed as a hairpin to illustrate their complementarity before bisulfite deamination of all unmethylated C residues. mC, 5-methylcytosine. The recognition site for BsmAI is in italics and its cleavage specificity is shown on the right. (B) Examples of discrimination between different methylated configurations of CpG dyads and C-to-T changes in a genomic sequence. Part of one strand of each molecular clone is depicted in the hairpin configuration to align CpG positions that were part of a genomic dyad.
