Figure 2.
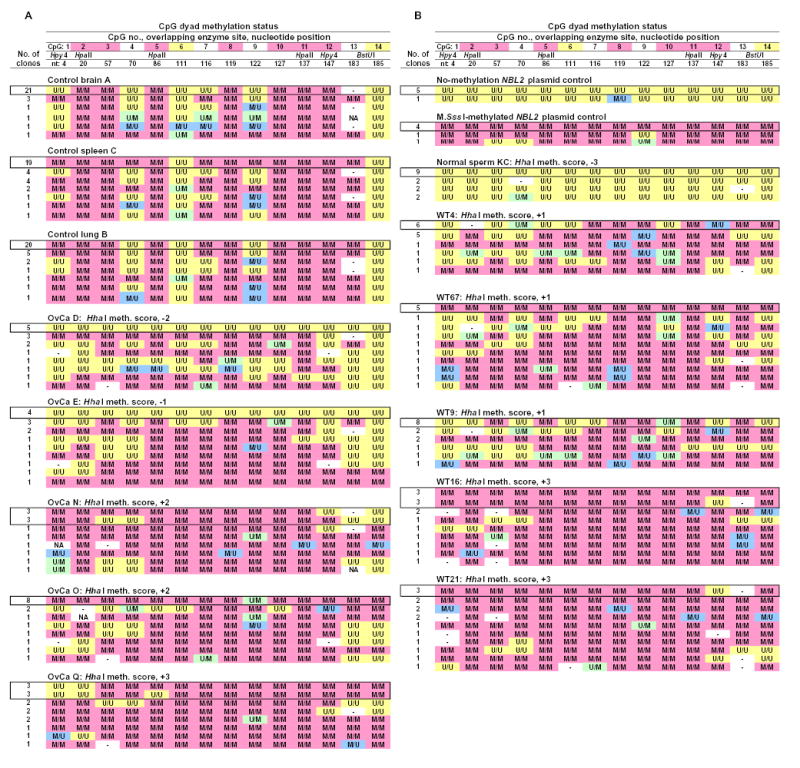
Hairpin genomic sequencing results from NBL2 for normal tissues, cancers, and in vitro-methylated (at CpG’s) or unmethylated NBL2 plasmid. Hairpin-bisulfite PCR-derived genomic sequences are shown for each clone. Each observed epigenetic pattern for a given sample illustrated only once. The most abundant pattern for each sample is boxed. M/M, a symmetrically methylated CpG dyad; U/U, a symmetrically unmethylated CpG dyad; M/U and U/M, the two orientations of hemimethylated CpG dyads; -, no CpG was present at that site due to sequence variation; NA, methylation could not be analyzed due to sequencing mistakes. Upon alignment of the two halves of the strands of the clones, sequencing mistakes were rare. At the top of each column, the following are given: the CpG site number (pink highlighting for sites always M/M in somatic controls and yellow for sites never M/M in somatic controls), nucleotide position within the post-primer sequenced region, and restriction sites. Hpy4, HpyCH4IV.
