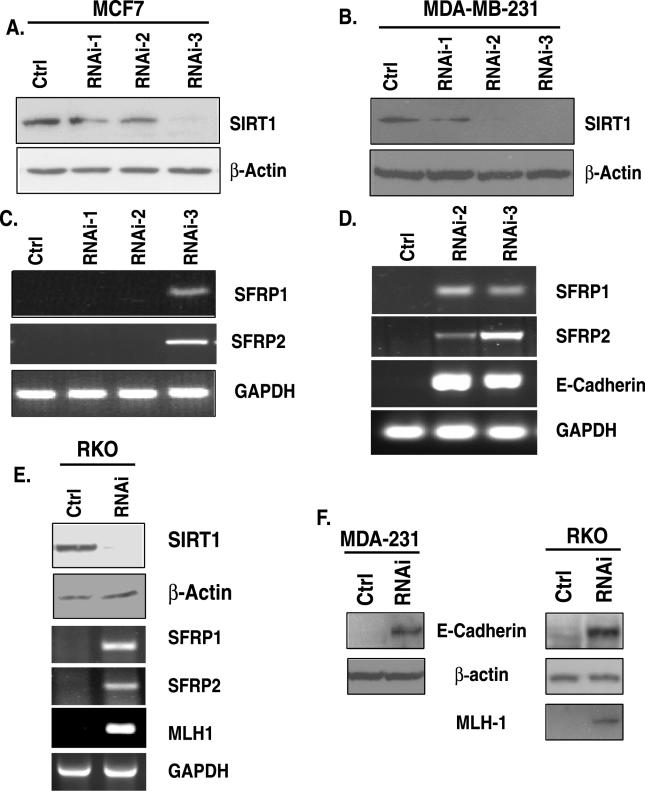
Figure 1. siRNA Knockdown of SIRT1 Causes Re-Expression of Epigenetically Silenced TSGs.
(A) RNAi-3 is most effective for reduction of SIRT1 in MCF7 cells. Retroviral expression vectors encoding SIRT1 cDNA that produce short hairpin loop RNA targeting either distinct regions of SIRT1 mRNA (RNAi-1, −2, or −3) or a control (ctrl) were used to infect MCF7. Western blot analysis for SIRT1 and β-actin was performed 48 h after two rounds of infection.
(B) Both RNAi-2 and −3 are effective for reduction of SIRT1 protein in MDA-MB-231 cells as described in (A).
(C) SIRT1 inhibition leads to TSG re-expression in MCF7 cells. RNA was isolated from parallel samples analyzed in (A), and RT-PCR was performed with intron-spanning primers specific for the genes SFRP1 and SFRP2. GAPDH was also analyzed as a control. Only the shRNA (RNAi-3) that caused substantial reduction in SIRT1 protein leads to gene re-expression. Control samples in which no reverse transcriptase was added were analyzed separately, and all were negative for amplification of the indicated genes.
(D) SIRT1 inhibition leads to TSG re-expression in MDA-MB-231 cells. RT-PCR was performed for analysis of the genes SFRP1, SFRP2, and E-cadherin as described in (A). Only the shRNAs (RNAi-2 and −3) that caused substantial reduction in SIRT1 protein lead to gene re-expression
(E) SIRT1 inhibition leads to TSG re-expression in RKO cells. SIRT1 protein reduction by RNAi-3 (top panel) as described in (A) leads to gene re-expression of SFRP1, SFRP2, and MLH1 as described in (C)
(F) MDA-MB-231 and RKO cells infected with control or RNAi-3 shRNA as described in (A) were selected with puromycin for 3 d, and pooled colonies were harvested for Western blot analysis of protein re-expression that corresponded with the gene reactivation described in (D) and (E).