Abstract
Efforts to correlate genetic variations with phenotypic differences are intensifying due to the availability of high-density maps of single nucleotide polymorphisms (SNPs) and the development of high throughput scoring methods. These recent advances have led to an increased interest for improved multiplex preparations of genetic material to facilitate such whole genome analyses. Here we propose a strategy for the parallel amplification of polymorphic loci based on a reduced set of nucleotides. The technique denoted Tri-nucleotide Threading (TnT), allows SNPs to be amplified via controlled linear amplification followed by complete removal of the target material and subsequent amplification with a pair of universal primers. A dedicated software tool was developed for this purpose and variable positions in genes associated with different forms of cancer were analyzed using sub-nanogram amounts of starting material. The amplified fragments were then successfully scored using a microarray-based PrASE technique. The results of this study, in which 75 SNPs were analyzed, show that the TnT technique circumvents potential problems associated with multiplex amplification of SNPs from minute amounts of material. The technique is specific, sensitive and can be readily adapted to equipment and genotyping techniques used in other research laboratories without requiring changes to the preferred typing method.
INTRODUCTION
Following the complete sequencing of the human genome, efforts have intensified to decipher the genetic code and ascertain the relationships between genetic variations and phenotypic differences, e.g. variations in disease susceptibility, drug responses and cancer ontogeny. Such analyses of genetic variations also have applications in diverse fields, such as genealogy, epidemiology and forensics. In some cases single nucleotide variants have been demonstrated to be associated with specific disease states, but in the vast majority of cases it is unlikely that a single polymorphism can be causally linked to a certain trait. Instead, certain combinations of variants among the millions of single nucleotide polymorphisms (SNPs) in the genome are likely to give rise to traits, such as increased risk for cancer. Consequently, accurate and sensitive analysis of SNPs may play an important role in future DNA diagnostics. Whole genome SNP typing platforms are already available to enable such high throughput scoring (1).
To facilitate these efforts, equally efficient methods for target amplification are needed. Padlock probes and linker ligated genomic tags, accompanied by amplification with universal probes, have demonstrated utility for such purposes (2–4), while use of multiplex PCR (i.e. PCR with multiple specific primers) has limitations since only a few loci can be amplified in parallel. Previous works have demonstrated that multiplex PCR with > 20 genomic fragments is problematic, in both theory and practice, due to its high complexity and complications associated with multiple, unspecific, competing annealing and extension events (5,6). These problems are circumvented in the approach presented here by reducing the number of nucleotides included in the extension reactions from four to three, leading to a predefined termination close to the extending probe. The central objective of this work is to provide a system for the simultaneous preparation and amplification of minute amounts of genomic DNA at a large number of genetic loci by a specific and sensitive tri-nucleotide threading procedure (TnT) that can be applied to large-scale scoring experiments in a single automated reaction sequence (see Figure 1).
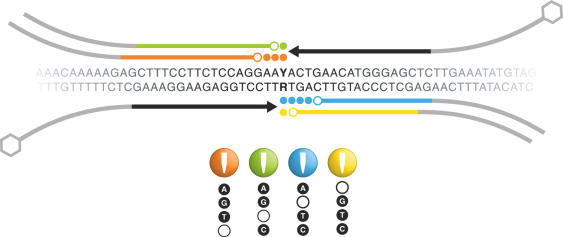
Figure 1.
Principle of the TnT technique. Illustration of the possible threads formed by different combinations of nucleotides for a specific polymorphic site, in the example above Y/R. Arrowheads indicate the 3′ ends of the extension primers. Nucleotides incorporated by the polymerase are illustrated by filled circles. The nucleotide at the 5′ end of each thread-joining primer is represented by an unfilled circle and is consequently the fourth non-present dXTP in the extension reaction. Ligation is carried out between a filled and an unfilled circle. Amplification tags are marked in gray and the hexagons denote biotin. See text for more details.
Overall, the TnT approach involves the following steps: (i) database searching and filtering; (ii) polymerase-mediated extension with just three dNTPs (tri-nucleotides); (iii) ligation of the 3′ end of the extended probe to the 5′ end of a second probe (thread-joining primer), which increases the specificity of the assay; (iv) cycling of the extension-ligation steps leading to linear amplification; (v) capture of the resulting ligated products (the DNA threads), resulting in complete removal of the genomic DNA; (vi) amplification of the ligated products by a pair of universal PCR primers and (vii) genotyping of the amplified fragments. In this study, the genotyping of the amplicons was carried out using a microarray-based variant of an allele-specific extension assay denoted protease-mediated allele-specific extension (PrASE) (7). However, since TnT represents a general solution to parallel amplification of genomic DNA, any other typing method capable of analyzing a single reaction harboring several different DNA fragments can be employed.
MATERIALS AND METHODS
SNP selection
The SNPs were identified in genes linked to various types of human cancers using the dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), Ensembl (www.ensembl.org) and SNP500Cancer (http://snp500cancer.nci.nih.gov) databases. This was followed by a filtering procedure, which primarily involved manual selection of candidate SNPs with relatively high allele frequencies in the Caucasian population and removal of SNPs with neighboring polymorphisms (<40 residues). The sequences were automatically retrieved in Fasta format from dbSNP (build 121) using a Perl script. The script also extracted the sequences 40 bases upstream and 40 bases downstream of each SNP. Note that SNPs with shorter surrounding sequences were removed. Subsequently, the human genome sequence was screened with the candidate SNP sequences using blastn (Genome build 35) to remove SNP sequences spanning duplicated gene segments or repetitive regions as well as low-complexity regions (see the SNP selection box in Figure 2).
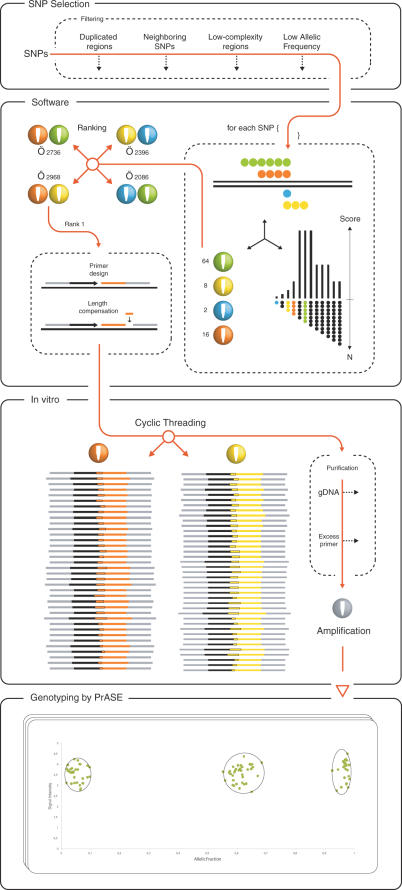
Figure 2.
Schematic diagram of the TnT technique for parallel genotyping. SNP selection: SNPs are selected and screened. Polymorphic regions with undesired features are filtered out. Software: each SNP is extended in silico and the different possible extensions are scored according to an exponential function. The SNPs are subsequently sorted to each of the four possible pairs of tri-nucleotide mixtures and the extension score is added to the combinations' total score. The combinations are then ranked according to their total scores and two primers, the extension primer and the thread-joining primer, are designed for each SNP. The length of the primers is adjusted so that the differences in the lengths of the threads do not exceed 10% (Length compensation). In vitro: the mixture of primers, enzymes, nucleotides and DNA template are subjected to cyclic extension-ligation reactions followed by an automated purification step to remove genomic DNA and excess primer. The two sets of reaction products are merged and the threads are amplified with a general primer pair. Genotyping: the different threads are decoded by a genotyping method, in this case PrASE, and allelic fractions are calculated for correct genotype scoring.
Algorithm and scoring
A software tool implemented in Java was designed to rank the possible tri-nucleotide combinations (AGT, AGC, ATC and GTC) and subsequently suggests two optimal tri-nucleotide reactions for a given set of SNPs (see Figure 2 the Software box and Supplementary Figure 1a).
The algorithm for ranking different tri-nucleotide combinations consists of the following steps (see also the Software box in Figure 2). (i) Extend the first SNP in silico with all possible tri-nucleotide sets for the SNP under study. The four possible tri-nucleotide sets (ACG/ACT, ACG/AGT, ACT/CGT and AGT/CGT) can be used for R, Y, K and M, while for W and S only two sets (AGT/TCA CTG/GAC) can be employed (Supplementary Figure 1b). (ii) Assign scores of 2, 4, 8, 16, 64, 64, 64, 32, 32, 32, 16 and 16 to 1-, 2-, 3-, 4-, 5-, 6-, 7-, 8-, 9-, 10-, 11- and 12-base extensions, respectively. (iii) Allocate the SNP to each of the possible tri-nucleotide reaction combinations so that each combination's total score is maximized. (iv) Repeat steps (i) to (iii) for all SNPs in the dataset. The tri-nucleotide combination with the highest score (rank) is the optimal choice. (v) The user confirms the selection of the combination with the highest score, or a preferred combination, and the software designs primers given amplification tags and a user-defined Tm window (see Supplementary Figure 1a). Note, in this study, the defined cutoff was 75 SNPs, and since the number of input SNPs was 88, the SNPs included in a certain combination may differ from those in another. However, the software tool is flexible, so the number of input SNPs may be exactly 75 or any number exceeding 75. The optimal tri-nucleotide combination for this dataset was dCTP, dTTP, dGTP (39 SNPs) and dATP, dGTP, dTTP (36 SNPs) with a total score of 2968. The 75 SNPs were located in 61 different genes, distributed amongst all but four chromosomes and included all of the different single nucleotide alterations (IUPAC codes: K, S, R, Y, W, M), as listed in Supplementary Table 1. (vi) The lengths of the final primers are adjusted to avoid PCR bias. Since the longest fragments were 115 bases long in this case, none of the DNA threads was allowed to be shorter than 104 bases (giving a maximum difference of 10%). The sequences of the primers are listed in Supplementary Table 2. (vii) In addition, the software designs genotyping primers. Given a set of signature tags, the software designs 3′-termini allele-specific primers (fused with signature tags at the 5′ end) for genotyping by PrASE (Supplementary Table 3).
Primers and template
All oligonucleotides were synthesized by MWG-Biotech AG (Ebersberg, Germany). For each primer pair (later forming a DNA thread) the extension primer was biotinylated at the 5′ end and the other was phosphorylated at the 5′ end. The signature tags of the generic tag arrays (anti-tags) were ordered with an amino group at the 5′-terminus for the array preparation (2) (Supplementary Table 4).
The genomic DNA used was an ECACC Human Random Control DNA Panel (SIGMA) isolated from lymphoblastoid cells from UK population with Caucasian origins (96 samples with DNA concentrations of 8 ± 2 ng/µl, treated according to the manufacturer's instructions). In addition, genomic DNA was isolated from blood samples from ten healthy anonymous Swedish Caucasian individuals.
TnT
Two reaction mixtures containing 0.2 mM of the dXTPs ‘C, T and G’ and ‘A, T and G’, 0.01 µM of each extension primer, 0.05 µM of each thread-joining primer and 8 ng genomic DNA were prepared in 20 mM Tris–HCl (pH 8.3), 25 mM KCl, 10 mM MgCl2, 0.5 mM NAD and 0.01% Triton X-100 together with 2 U Ampligase (Epicentre) and 0.5 U Stoffel fragment DNA polymerase (Applied Biosystems). For each set of ten individuals to be analyzed, one reaction without genomic DNA and one reaction without ligase were included as negative controls.
The reactions were performed in a GeneAmp 9700 PCR System (Applied Biosystems) with a temperature profile consisting of an initial pre-cycling step (20°C for 5 min, 95°C for 5 min) followed by 99 annealing, extension and ligation cycles (95°C for 15 s and 65°C for 12 min).
Thread clean-up
In order to remove genomic DNA and excess primer, a cleansing procedure was set up robotically with a Magnatrix 1200 (Magnetic Biosolutions) liquid handling robot running custom made scripts. Prior to immobilization of the DNA threads on to streptavidin-coated Dynabeads DP (Dynal), each reaction tube was subjected to Proteinase K treatment (20 µg) for 20 min at 60°C and the contents of the two tri-nucleotide reaction tubes were pooled. After immobilization of the DNA threads, the beads were magnetically collected and washed with 1× TE [10 mM Tris–HCl (pH 7.5), 1 mM EDTA] followed by thorough washes with water (Millipore) and 0.1 M NaOH with constant mixing. Finally, the DNA threads were released by heating to 80°C for 1 s in water (Magnetic Biosolutions) (8).
Parallel thread amplification
Following release, the 75 DNA threads were amplified in a single PCR tube. The PCR mixture consisted of 10 mM Tris–HCl (pH 8.3), 2 mM MgCl2, 50 mM KCl, 0.1% (v/v) Tween-20, 0.2 mM dNTPs, 0.2 µM of each universal primer and 1 U of AmpliTaq Gold DNA polymerase (Applied Biosystems) in a total volume of 50 µl. One of the amplification primers was biotinylated. After activating the polymerase at 95°C for 12 min the reaction was cycled 60 times using a profile consisting of 95°C for 30 s, 68°C for 30 s and 72°C for 30 s, followed by a final elongation step at 72°C for 2 min.
Array preparation, PrASE-genotyping and data analysis
Generic signature tag microarrays (with 16 sub-arrays per array) were designed and prepared using a QArray system (Genetix). Each of the 16 sub-arrays printed on a slide accommodated a duplicate of 150 tags, allowing the analysis of 75 SNPs by PrASE-genotyping (7). In brief, PrASE is an alternative approach to conventional allele-specific primer extension (ASE) assays but has increased specificity features. The ASE assay uses the inherent ability of DNA polymerases to extend 3′-termini mismatched primers discriminately. However, certain 3′-termini mismatched sequences cannot be distinguished by the DNA polymerases, resulting in comparable extensions of matched and mismatched primer complexes. This problem can be circumvented by exploiting the fact that mismatched primers have slower reaction kinetics, by adding a protease in the extension reaction.
Following the PrASE reaction, data were obtained by scanning the slide with an Agilent scanner (Agilent Technologies) and were analyzed using GenePix Pro 5.1 software (Axon Instruments). The median local background intensities were subtracted from the median intensities of the spots by GenePix Pro 5.1 and the data were analyzed in Microsoft Excel, where the mean values of fluorescence intensities of the duplicates for each signature tag were used to calculate the allelic fractions of the 75 SNPs for each individual. Allelic fractions were calculated as intensity of spot A/(intensity of spot A + intensity of spot B) and were plotted against the logarithm of the total signal intensities of spot A + spot B (see Figure 4).
RESULTS AND DISCUSSION
The TnT assay analyzes all six classes of SNPs (substitution polymorphisms): K (G/T), M (C/A), Y (C/T), R (G/A), W (A/T) and S (C/G). In order to analyze all of these SNPs by tri-nucleotide extensions, four tri-nucleotides (AGT, TCA, CTG and GAC) may be required. For example, a K polymorphism may be extended by nucleotide combinations AGT or CTG. Since the polymorphism can be scored on either strand, the complimentarity of the tri-nucleotide mixtures can be exploited to reduce the number of extension reactions required to cover all genetic variants, from four to two.
Figure 1 illustrates four extension scenarios for a polymorphic position with a C to T base variation. As shown in the figure, all four tri-nucleotide combinations may be employed to extend over this SNP. When the DNA strand with the C/T (Y) polymorphism is used as the target template, the combinations AGT (indicated by red) or AGC (indicated by green) can be used to generate a complementary strand—a DNA thread. Note that the nucleotides G and A have to be present in order to extend over and cover both allelic variants. When the AGT nucleotide combination is used in the reaction, the extension primer is extended by 3 nt when either allele variant provides the template. In this case, if the polymorphic position in the target template is homozygous C/C, the nucleotides ‘gtt’ will be incorporated, and since the nucleotide C is not included, the extension will be terminated when C has to be incorporated and the extended product can be ligated to a downstream probe (thread-joining primer). Similarly, the extension will be terminated at the same position with both homozygous and heterozygous variants. However, if the polymorphic position in the target template is homozygous T/T the polymerase will incorporate ‘att’ nucleotides, and when the sample is heterozygous C/T, the target alleles with the C and T variants will induce the incorporation of ‘gtt’ and ‘att’, respectively. Thus, both allele products can be ligated to the second primer and amplified in the PCR that follows. Accordingly, when the AGC nucleotide combination is used, only one base can be incorporated into the extension primer. As shown in Figure 1, the extension primer in both chosen cases (AGT or AGC nucleotide combinations) is the same and its 3′ end is situated next to the polymorphic site. However, the thread-joining primer has to be relocated depending on the number of incorporated bases in the gap.
As illustrated in Figure 1 and described above, the user of the technique can select any of the four tri-nucleotide sets. However, in order to compensate for possible unspecific extension/ligation events, an extension-length scoring system has been implemented, favoring extensions with more than 1 nt. After a cyclic extension/ligation reaction and complete removal of genomic DNA, the products are subjected to PCR amplification, using a pair of universal primers, followed by microarray-based PrASE to score the SNPs. Thus, the genotyping primer functions not only as a scoring probe but also as an additional specificity factor. Note that almost all steps in this method are automated by using a robotic workstation capable of handling magnetic beads.
The bioinformatic steps involve screening the human genome with the candidate SNP sequences to remove SNPs with sequences spanning duplicated gene segments sharing high sequence similarities, repetitive regions, low-complexity sequences and those with neighboring polymorphisms (see the SNP selection box in Figure 2). In addition, we developed a software tool to make in silico extensions with the four different tri-nucleotide combinations. An exponential extension-length scoring system that awards high scores to the most theoretically favorable extension-lengths is implemented in the software. Since two reactions have to be conducted to produce the DNA threads, the software calculates the total score for two tri-nucleotide combinations (AGC/CTA, AGC/AGT, CTG/CTA and CTG/AGT) and selects the combination with the highest score, then automatically designs primers (with desired Tm) for the multiplex reaction and the PrASE reaction (see the Software box in Figure 2 and Supplementary Figure 1). The extension-length scoring criteria have been set to favor long extensions (but not too long, see below), since formation of some unspecific DNA threads may be unavoidable even though the TnT technique should prevent the extension step entering an exponential phase and subsequent specific joining. However, if an unspecific event takes place, the likelihood that the unwanted thread will be discriminated by the genotyping primer(s) is dramatically increased if the number of nucleotides incorporated in the specific threads is raised. Consequently, DNA threads formed with a single nucleotide incorporation are given the lowest score (2) and extensions spanning 5–7 bases the highest score (64). The reason for implementing an exponential scoring system was to clearly differentiate between the different extensions lengths. However, to avoid PCR bias associated with product length, long extensions, such as 11 and 12 are given moderate scores (16). In addition, the software adjusts the length of the primers of short DNA threads, ensuring that the differences in length between the longest and shortest threads do not exceed 10% (see the Software box in Figure 2). Since the majority of the steps in the TnT technique are performed automatically using a bead-handling robotic workstation, a custom made script dedicated to the project was developed.
After a successful pilot experiment with the TnT approach, comparing array based and Pyroseqeuncing genotyping technology (data not shown) we chose to proceed with the array platform. Hence, SNPs located in genes related to different forms of cancer were identified and randomly selected using the dbSNP, SNP500Cancer- and Ensembl databases. Our current array of generic tag arrays has the capacity to genotype 75 SNPs (using the PrASE technique, which requires two allele-specific primers/SNP), so the software was designed to select 75 SNPs. The nucleotide mixture selection was based on the scoring system where the tri-nucleotide combinations of AGT and CTG received the highest total score of 2968. Using this combination, 39 SNPs would be in the CTG set and 36 in the AGT set. The scores (and SNP distributions) of combinations AGC/AGT, AGC/CTA and CTG/CTA were 2736 (36/39), 2086 (46/29) and 2396 (54/21), respectively. The user has the option to make a final decision to select a more suitable combination. Such a scenario may occur when two or more combinations have obtained close scores and the software selects the combination with the highest score, although the SNP distribution suggests that one of the other combinations would be better. Nevertheless, the 75 SNPs selected in this study (combinations of AGT/CTG) are represented on all chromosomes except chromosomes 12, 21; X and Y (see Supplementary Table 1).
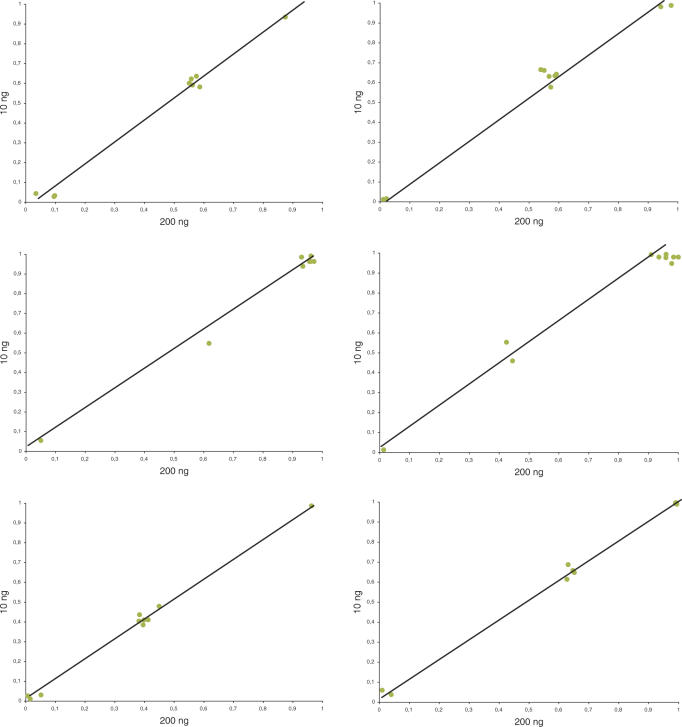
Genomic DNA from a panel of ten Swedish individuals was subjected, at two concentrations (200 and 10 ng/µl), to 75-plex amplification using the TnT approach, and the individuals were genotyped using the PrASE protocol. Genotyping results obtained with the two concentrations of genomic DNA showed complete correlation, indicating that the approach can be used for parallel amplification of very small amounts of target genomic DNA as starting material (Figure 3).
Figure 3.
TnT results related to concentration. Correlation in genotype using 200 ng (x-axis) and 10 ng (y-axis) DNA for the SNPs 20, 36, 48, 55, 61 and 63.
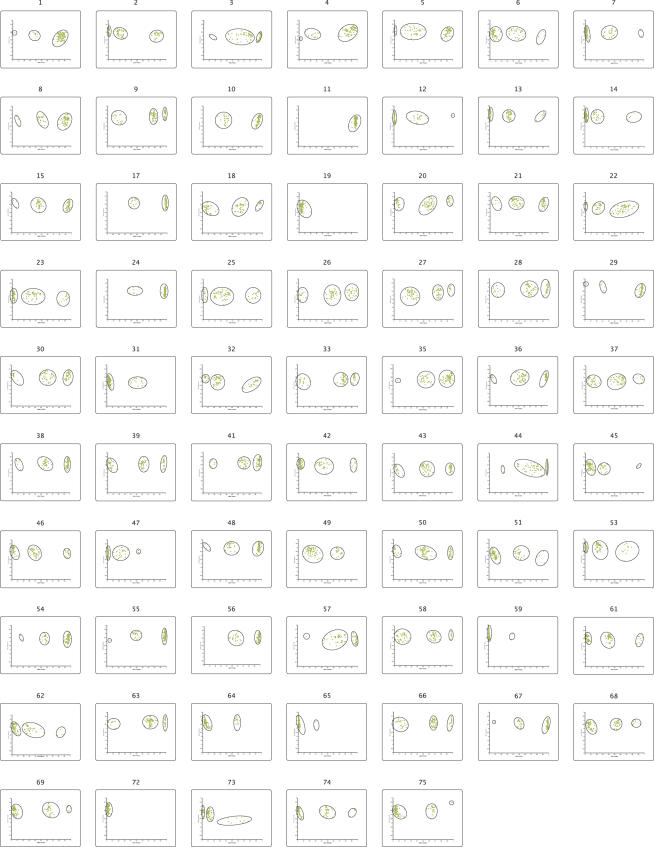
Consequently, the DNA threading was carried out using sub-nanogram amounts of genomic DNA as starting material. DNA from a total of 96 Caucasian individuals (UK population) was then amplified with the TnT protocol and genotyped with PrASE. 68 out of the 75 assayed SNPs (93%) rendered partitioned clusters of homozygous and heterozygous genotypes (Figure 4). The SNPs that did not give satisfactory clusters (mostly due to very weak signals) were SNPs 16, 34, 40, 52, 60, 70 and 71. The SNPs 52 and 60 did however give relatively good signals but cluster partitioning was not satisfying.
Figure 4.
Cluster diagrams. Cluster diagrams for the 68 SNPs analyzed.
To investigate the cause of the weak signals for the remaining SNPs, these were subjected to the TnT protocol, but were amplified and genotyped in a duplex fashion (by including a control SNP as a reaction control). Despite the low-complexity in terms of multiplexing, the array signals from SNPs 34, 40 and 70 remained at background level and did not result in correct clustering. This may have been due to failures in oligonucleotide synthesis or errors in the databases. The genotyping of SNPs 16 and 71 resulted in relatively strong signals and generated clusters. In conclusion, 68 out of 72 possible SNPs gave partitioned clusters (conversion rate > 94%) and 107 out of the 6528 (68*96) possible genotypes did not fall into the clusters (call rate > 98%). The typing results for each SNP (the allele frequencies) were compared to the allele frequencies given by dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). Considering that the data in dbSNP provides the frequencies of several different populations while the data in this work reflects frequencies of a very homogeneous population (UK Caucasians), the typing results (resulted from TnT) are in very good agreements with dbSNP's (see Supplementary Table 5).
In addition, in order to investigate the sensitivity of the approach, a series of dilutions of two samples was prepared including 8, 4, 2, 1 and 0.5 ng of genomic DNA, which was subjected to parallel DNA threading. The genotyping results were then included in the cluster diagrams obtained from the analysis of the 96 samples with 8 ng of genomic DNA. Identical genotyping scores were obtained for all 68 of the working SNPs in concentrations down to 1 ng per reaction, corresponding to ∼150 cells. With 0.5 ng DNA, some of the SNPs did not give reliable scores due to weak signals.
To our knowledge, the TnT technique provides a unique means to amplify any given polymorphism in small amounts of starting material. A number of techniques, based on fragmentation of the genomic DNA and subsequent amplification (9–11) and direct typing on whole genome amplified DNA (12), have recently been described. In addition, a recent study demonstrated co-amplification of more than 1000 selected SNPs by general PCR (13). But all these techniques have a major disadvantage since they lack the ability to genotype any specified polymorphic position. There are however two techniques, the Golden Gate assay (Illumina Inc) (14) and the MIP assay (Parallele Biosciences) (15), that are capable of parallel amplification of specified polymorphisms. The Golden Gate assay is based on ASE followed by ligation, but unspecific 3′ end extensions of certain mismatches (16) tend to occur, and the technique requires careful optimizations. In addition, unspecific primer hybridization in combination with uncontrolled extensions, due to the presence of all dNTPs, makes it impossible to initiate linear amplification so relatively large amounts of starting material, 0.2 to 1 µg of genomic DNA, are required. The MIP assay uses specific pre-circle probes that are locked (padlocked) to the target molecule after gap-filling and ligation steps. However, the padlock formation may prevent cycling of the gap-fill/ligation procedure and consequently 0.4 to 2 µg of genomic DNA may be required in the assay, while the TnT technique provides reliable results with as little as 1–9 ng (100–1000 cells) of genomic DNA without employing biased whole genome amplification (17,18). An apparent advantage of the Golden Gate and MIP assays is that they integrate amplification with the genotyping, while the TnT approach focuses solely on amplification. However, despite the need to add a genotyping step, this may be advantageous because the technique can be applied in any genotyping laboratory without changing the typing routines. In addition, as noted above, the genotyping primers in combination with the incorporation of more than one base in the formation of the DNA threads gives high specificity, as reflected in the cluster diagrams and the fact that 68 out of 72 SNPs worked in the assay. A possible drawback that the TnT technique shares with most genotyping and amplification techniques, in particular with the Golden Gate and MIP assays, is the negative effect of neighboring SNPs on the assay performances. As some SNPs have a neighboring sequence variation with a distance closer than 40 bases, these could affect the probe hybridization efficiency and are recommended to be avoided.
In conclusion, we present a specific and sensitive technique for the multiplex amplification of DNA sequences with variable positions (SNPs). A 75-plex approach may represent a high degree of complexity and cost (approximately USD 0.056/SNP), for academic facilities with modest financial resources, but the TnT technique has great potential to specifically amplify thousands of variable segments. The sensitivity of analytical techniques is a very important issue, as well as their specificity, when the numbers of cells available is limited, as they often are in analyses of tumor materials, forensic investigations and attempts to identify biomarkers using biobanks for instance (19). Since the TnT technique can give identical results with 1, 8 or 200 ng of genomic DNA, we believe that by taking advantage of signal amplification strategies (12,20) it can retain much of its sensitivity when a 1500-plex approach is designed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Research Council and the Knut and Alice Wallenberg Foundation (Wallenberg Consortium North). Funding to pay the Open Access publication charges for this article was provided by Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang D.G., Fan J.B., Siao C.J., Berno A., Young P., Sapolsky R., Ghandour G., Perkins N., Winchester E., Spencer J., et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 2.Hirschhorn J.N., Sklar P., Lindblad-Toh K., Lim Y.M., Ruiz-Gutierrez M., Bolk S., Langhorst B., Schaffner S., Winchester E., Lander E.S. SBE-TAGS: an array-based method for efficient single-nucleotide polymorphism genotyping. Proc. Natl Acad. Sci. USA. 2000;97:12164–12169. doi: 10.1073/pnas.210394597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsson M., Malmgren H., Samiotaki M., Kwiatkowski M., Chowdhary B.P., Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 4.O'Meara D., Ahmadian A., Odeberg J., Lundeberg J. SNP typing by apyrase-mediated allele-specific primer extension on DNA microarrays. Nucleic Acids Res. 2002;30:e75. doi: 10.1093/nar/gnf074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsson M., Baner J., Mendel-Hartvig M., Dahl F., Antson D.O., Gullberg M., Landegren U. Making ends meet in genetic analysis using padlock probes. Hum. Mutat. 2002;19:410–415. doi: 10.1002/humu.10073. [DOI] [PubMed] [Google Scholar]
- 6.Landegren U., Nilsson M. Locked on target: strategies for future gene diagnostics. Ann. Med. 1997;29:585–590. doi: 10.3109/07853899709007487. [DOI] [PubMed] [Google Scholar]
- 7.Hultin E., Kaller M., Ahmadian A., Lundeberg J. Competitive enzymatic reaction to control allele-specific extensions. Nucleic Acids Res. 2005;33:e48. doi: 10.1093/nar/gni048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmberg A., Blomstergren A., Nord O., Lukacs M., Lundeberg J., Uhlen M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy G.C., Matsuzaki H., Dong S., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., et al. Large-scale genotyping of complex DNA. Nature Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- 10.Dahl F., Gullberg M., Stenberg J., Landegren U., Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunderson K.L., Steemers F.J., Lee G., Mendoza L.G., Chee M.S. A genome-wide scalable SNP genotyping assay using microarray technology. Nature Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 13.Wang H.Y., Luo M., Tereshchenko I.V., Frikker D.M., Cui X., Li J.Y., Hu G., Chu Y., Azaro M.A., Lin Y., et al. A genotyping system capable of simultaneously analyzing >1000 single nucleotide polymorphisms in a haploid genome. Genome Res. 2005;15:276–283. doi: 10.1101/gr.2885205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J.B., Oliphant A., Shen R., Kermani B.G., Garcia F., Gunderson K.L., Hansen M., Steemers F., Butler S.L., Deloukas P., et al. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 15.Hardenbol P., Baner J., Jain M., Nilsson M., Namsaraev E.A., Karlin-Neumann G.A., Fakhrai-Rad H., Ronaghi M., Willis T.D., Landegren U., et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 16.Newton C.R., Graham A., Heptinstall L.E., Powell S.J., Summers C., Kalsheker N., Smith J.C., Markham A.F. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergen A.W., Qi Y., Haque K.A., Welch R.A., Chanock S.J. Effects of DNA mass on multiple displacement whole genome amplification and genotyping performance. BMC Biotechnol. 2005;5:24. doi: 10.1186/1472-6750-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergen A.W., Haque K.A., Qi Y., Beerman M.B., Garcia-Closas M., Rothman N., Chanock S.J. Comparison of yield and genotyping performance of multiple displacement amplification and OmniPlex whole genome amplified DNA generated from multiple DNA sources. Hum. Mutat. 2005;26:262–270. doi: 10.1002/humu.20213. [DOI] [PubMed] [Google Scholar]
- 19.Maschke K.J. Navigating an ethical patchwork–human gene banks. Nat. Biotechnol. 2005;23:539–545. doi: 10.1038/nbt0505-539. [DOI] [PubMed] [Google Scholar]
- 20.Steemers F.J., Chang W., Lee G., Barker D.L., Shen R., Gunderson K.L. Whole-genome genotyping with the single-base extension assay. Nature Meth. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.