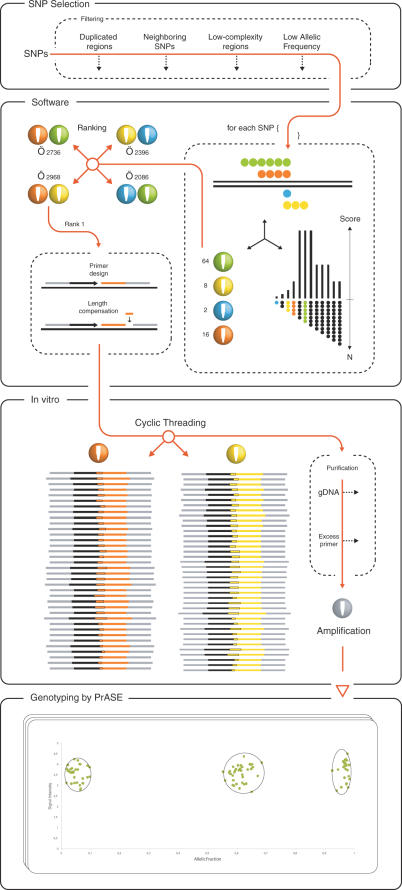
Figure 2.
Schematic diagram of the TnT technique for parallel genotyping. SNP selection: SNPs are selected and screened. Polymorphic regions with undesired features are filtered out. Software: each SNP is extended in silico and the different possible extensions are scored according to an exponential function. The SNPs are subsequently sorted to each of the four possible pairs of tri-nucleotide mixtures and the extension score is added to the combinations' total score. The combinations are then ranked according to their total scores and two primers, the extension primer and the thread-joining primer, are designed for each SNP. The length of the primers is adjusted so that the differences in the lengths of the threads do not exceed 10% (Length compensation). In vitro: the mixture of primers, enzymes, nucleotides and DNA template are subjected to cyclic extension-ligation reactions followed by an automated purification step to remove genomic DNA and excess primer. The two sets of reaction products are merged and the threads are amplified with a general primer pair. Genotyping: the different threads are decoded by a genotyping method, in this case PrASE, and allelic fractions are calculated for correct genotype scoring.