Abstract
Objective
To investigate the relationship between number and location of allelic imbalances (AI) and local tumor progression according to Astler-Coller classification.
Summary Background Data
Spontaneous errors in DNA replication (i.e., allelic imbalance or microsatellite instability) have been suggested to play an important role in carcinomatous transformation as reflecting alterations of gene function.
Methods
One hundred two consecutive patients with colorectal carcinoma undergoing surgical resection were included in this study. Patients were distributed according to the Astler-Coller classification as stages A (n = 7), B1 (n = 15), B2 (n = 24), C (n = 31), and D (n = 25). Fluorescent polymerase chain reaction was performed on frozen tumor, normal colon mucosa, and blood DNA at 35 microsatellite markers. Allelic imbalance frequency was compared with tumor staging.
Results
The percentage of AI was significantly higher in stage D than in A/B1 and B2. In addition, the percentage of AI was significantly higher in 10 synchronous colorectal liver metastases than in stage A/B1 and B2 tumors. However, the allelotyping revealed a subgroup of A/B1 tumors with a high AI frequency. Statistical analysis showed that the presence of AI at microsatellites D1S305, D2S138, D3S1282, D17S790, and D22S928 presented a significantly positive correlation with stages.
Conclusion
The frequency of AI significantly correlates with tumor progression of colorectal cancer. Primary tumors with synchronous colorectal liver metastases showed a higher percentage of AI, suggesting that a frequency of AI greater than 35% with this selection of markers indicates a high risk of local progression and of development of metastases.
Colorectal carcinoma is the second most common cause of cancer deaths in the Western world, and death is mainly due to metastatic liver involvement. Colorectal carcinoma provides an excellent opportunity to study the adenoma-to-carcinoma sequence, the progression of the stage of the disease, and metastatic events. During the past decade, since the original results of allelotyping performed by Vogelstein et al, 1 numerous molecular and cytogenetic studies have strengthened the hypothesis that stepwise accumulation of defective tumor suppressor genes or mutated oncogenes, or both, is involved in the genesis and the progression of colorectal carcinoma. 2–6 Sequential alterations were supposed to appear during the development of the tumor from adenoma to carcinoma, and then metastases including alterations at multiple chromosome arms such as losses of 5q, then losses of 18q followed by 17p losses involving genes such as APC, DCC, and p53. 7 Other molecular alterations were observed identifying potential chromosome regions such as 8p, 9q, 10p, 13q, 11q, 19q and other mutated genes such as β-catenine or c-MET. 8–17
Microsatellites are highly polymorphic repeated sequences (mono- to tetranucleotides repeated sequentially 20 times on average) localized randomly mostly in noncoding regions. Modifications or alterations at these repeated sequences have been shown to occur in human tumors. 1 These genomic rearrangements in microsatellites can be broadly divided into two main groups: microsatellite instability (MSI) and allelic imbalance (AI). MSI describes the accumulation of mutations or modifications in the number of repeats resulting from failure of the DNA mismatch repair mechanism. Tumors that display MSI frequency up to 30% are described as RER+ (Replication ERror) or MSI-H. 17–22 Several studies have shown that RER is found in approximately 90% of patients with hereditary nonpolyposis colorectal (HNPCC) cancer and in 15% to 20% of those with sporadic colorectal carcinomas. 17–19 Accordingly, other studies showed that RER tumors belong to a specific group with a better prognosis. 23,24 AI refers to the partial or complete loss of one of the two alleles (previously known as loss of heterozygosity [LOH]) or alternatively to the amplification of one allele compared with the other. Identification of LOH in tumor cells would mean the presence of tumor suppressor genes at these loci. 1,7 LOH tumors, which represent more than 80% of all colorectal cancers, are mainly classified in terms of histologic and pathologic features. Considering the increasing complexity of alterations described during tumor progression, molecular analysis could help us understand the mechanisms underlying the existence of recurrences and invasiveness of early-stage cancers.
To classify more precisely these tumors based on their molecular status, we decided to perform allelotyping with a sensitive and automated method using fluorescent-based DNA technology; this allows more precise quantification of the polymerase chain reaction (PCR) products than radioactive assays. 25
We used a panel of 35 microsatellites mainly localized in chromosome regions previously described as being frequently altered. We investigated, in 102 consecutive patients with colorectal carcinomas staged according to the Astler-Coller classification, the relationship between AI and local tumor progression or occurrence of synchronous colorectal liver metastases.
METHODS
Patients and Tumor Specimens
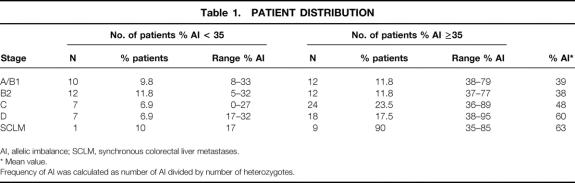
In our institution, from February 1995 to October 1998, 118 patients were analyzed. One hundred two consecutive patients (59 men, 43 women) undergoing resection of primary colorectal cancer were included. The mean age was 64.3 years (range 34–85). Patients were distributed in accord with the Astler-Coller classification as stage A (n = 7), stage B1 (n = 15), stage B2 (n = 24), stage C (n = 31), and stage D (n = 25) (Table 1). Among the 25 patients with stage D, 21 had synchronous colorectal liver metastases and 4 had peritoneal deposits. Liver metastases of 10 of the 21 patients with synchronous colorectal liver metastases were allelotyped. During the analysis, 16 patients were identified as MSI-H using at least five informative microsatellites in accord with the National Institutes of Health recommendations 20 and were not included in the population of 102 patients. The primary tumor was localized in 44 patients in the right colon, in 43 in the left colon, and in 15 in the rectum. The frozen tumors were microdissected at the Pathology Department of our institution.
Table 1. PATIENT DISTRIBUTION
AI, allelic imbalance; SCLM, synchronous colorectal liver metastases.
* Mean value.
Frequency of AI was calculated as number of AI divided by number of heterozygotes.
Tissue and Blood DNA Extraction
For each patient, DNA was extracted from blood, tumor sections containing at least 30% of tumor cells as estimated by the pathologist, and normal paired frozen mucosa sections using classical phenol-chloroform extraction, as previously described. 26
Microsatellite Markers, PCR, and Analysis
Extracted DNA from each sample was amplified by PCR using fluorescent primers, as described previously. 26–28 Thirty-five polymorphic microsatellite markers were analyzed, targeting 17 chromosome loci. This panel corresponds to frequently rearranged loci 29,30. Microsatellite markers and chromosomal locations are summarized in Table 2. All the primer sets were obtained through the Genome Data Base (www.gdb.org) or Genemap’98 (www.ncbi.nlm.nih.gov/genemap99). PCR amplification were carried out using the Taq polymerase (GIBCO-BRL Life Technology, Rockville, MD) on the thermocycler (Omnigen Hybaid) for 35 cycles as follows: 95°C for 1 minute, 50°C for 1 minute, 72°C for 1 minute, followed by a final 5 minutes of extension at 72°C, as previously described. 26 One primer of each couple was fluorescent, and the amplified fragments were analyzed on an ALF Sequencer (Amersham-Pharmacia, Freiburg, Germany). This technique allows a quantitative evaluation of the allele ratio by measuring the peak height of both alleles and greatly improves the sensitivity of analysis. The use of unique labeling allows accession to raw data. 26 An informative microsatellite corresponds to heterozygosity and thus to the presence of two peaks (Fig. 1, panel A). Allelic imbalance refers to partial or complete loss of one of the two alleles, resulting in a loss of heterozygosity, or alternatively amplification of one allele compared with the other, leading to a modification of the allele ratio in the tumor tissue compared with the allele ratio obtained in normal tissue or leukocytes (see Fig. 1, panel B). The presence of an AI was confirmed by at least two independent PCRs. Previous study allowed us to determine cutoff values for significant AI determined at 15%. 26 In the previous study, 26 the measurement of the peak height presented a good reproducibility and enabled us to determine the variability of the allele ratio between paired control or tumor tissue and blood DNAs. The intensity of AI was calculated as a percentage 26,28 : AI% = absolute value ([Bb/Ba] minus [Tb/Ta]) times 100/(Bb/Ba), in which Ba and Bb represent the height of the two alleles in the blood and Ta and Tb in the tumor tissue.
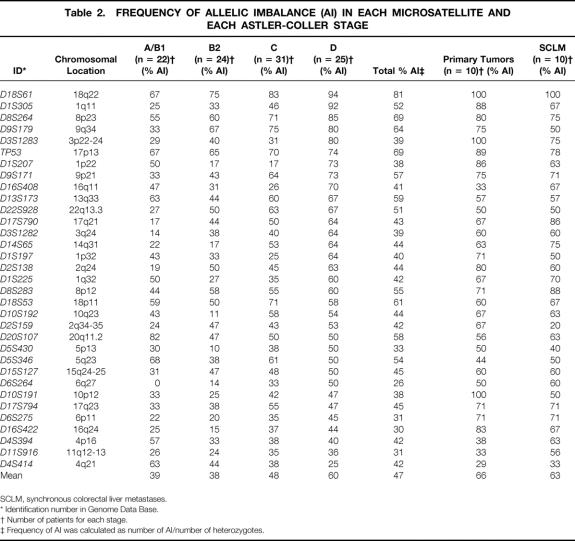
Table 2. FREQUENCY OF ALLELIC IMBALANCE (AI) IN EACH MICROSATELLITE AND EACH ASTLER-COLLER STAGE
SCLM, synchronous colorectal liver metastases.
* Identification number in Genome Data Base.
† Number of patients for each stage.
‡ Frequency of AI was calculated as number of AI/number of heterozygotes.
Figure 1. Electrophoresis examples of microsatellite amplification polymerase chain reaction (PCR). Genomic DNA was extracted from blood (B), normal tissue (N), primary tumor (T), and colorectal liver metastasis (M). Fluorescent PCRs of two microsatellites (A, D2S159; B, D8S283) were analyzed by using a sequence analyzer. The two heterozygote microsatellites were informative for allelic imbalance. (A) Normal pattern with no variation of the allele ratio in T and M samples versus B and N controls. (B) Allelic imbalance (AI) showing an alteration of the allele ratio in T and M samples (arrowheads) versus B and N controls.
In addition, for each patient and each microsatellite, we systematically amplified control leukocyte DNA and normal tissue DNA in parallel with the tumor DNA and determined the allele ratio of these two controls. In each case, the variations of allelic ratio between the two paired controls were always less than 15%. The panel of microsatellites contained dinucleotide repeats, excepted two mono-repeat markers (BAT26, TGFβRII). These two were used to check for RER phenotype and were excluded for further analysis of AI frequency.
Statistical Analysis
Each value was expressed as a mean ± standard error. Statistical analysis was performed with one-way factorial analysis of variance. When the F test showed a value to be significant, the Fisher test was used as a post-hoc multiple comparison. To compare the correlation between AI frequency of each microsatellite and Astler-Coller classification, we performed the chi-square test and the Fisher exact test. A difference was considered significant at P < .05.
RESULTS
The allelotyping was performed at 35 loci on 118 consecutive patients whose primary colorectal cancer were resected. Among them, 16 patients with MSI-H phenotype were characterized and excluded from further statistical analysis. Allelotyping was performed on liver metastases, which were resected or biopsied simultaneously during the same operative procedure for 10 of 102 patients. All our dinucleotide repeat microsatellites were chosen from the Genome Data Base and Genemap’98 depending on their percentage of heterozygosity above 70%, generally considered a good informative value. In our study, 26 microsatellites presented effectively a percentage of at least 70%. The seven other microsatellites (D9S179, D17S794, D4S414, D16S408, D351282, D5S430, D6S264) showed lower percentages (52%, 61%, 69%, 60%, 62%, 46%, and 64%, respectively) and thus appeared to be slightly less informative than usually reported.
Increase of AI Frequency in Accord With Astler-Coller Classification
In using the panel of 35 microsatellites, each tumor showed at least one alteration either as AI or as MSI, confirming the presence of tumor cells in the specimens. Thirty-five patients of the 102 showed, in addition to AI, one to three MSIs but were still considered as having LOH phenotype (MSI-L). 20 In stage A/B1, these sporadic MSIs were observed at only 4 microsatellites, whereas such alterations were detected at 12 to 15 microsatellites in further stages. The percentage of MSI in MSI-L patients ranged from 3% to 19%. Nevertheless, no significant increase in the number of MSIs per patient was observed through stages. Because there has been no explanation for the biologic meaning of such MSI at a noncoding region, only AI was considered in the statistical analysis.
The mean value of total AI frequency in the 102 specimens of colorectal carcinoma was 47% (0–95%). After stratification of the 102 patients according to the Astler-Coller classification (see Table 1), it was observed that the AI frequency was significantly higher in stage D colorectal tumor (60 ± 2.8%) than in stage A/B1 (39 ± 3.3%;P < .01) and stage B2 (38 ± 2.9%;P < .01) using one-way factorial analysis of variance and the Fisher test as a post-hoc comparison. In contrast, the statistical comparison between stage C versus stage D on the one hand and stage A/B1 versus B2 on the other revealed no significant difference. In Figure 2, stratification of the overall population based on their AI frequency shows that several tumors had a low percentage of alterations; further, a major group of patients had an AI frequency ranging from 30% to 40%. Thus, to characterize the different stages of tumors at the molecular level, patients were classified arbitrarily into two groups based on their tumor AI frequency (group 1, <35%; group 2, ≥35%) and in accord with the Astler-Coller classification. Using this limit, we observed that in stage A/B1 as well as stage B2, half of the patients belonged to group 2, in contrast to the distribution observed in stages C and D, where more than two thirds of the patients were classified as group 2. The statistical analysis comparing the distribution in stages A/B1-B2 versus C-D was significant (P = .016, chi-square). Seven stage A/B1 patients had an even higher AI frequency (>50%). With the threshold at 50%, the distribution of patients in the two groups was still significantly different when comparing stages A/B1-B2 and stages C-D (P = .034, chi-square).
Figure 2. Distribution of allelic imbalance (AI) frequency in overall population. For each colon tumor, the AI frequency was determined as a percentage of the number of AI versus the number of informative analyzed loci and classified into 10 groups. The number of tumors per group was expressed as a percentage.
The AI frequency was significantly higher in the patients with synchronous colorectal liver metastasis (63 ± 2.7%) than in the stage A/B1 (P < .05) and stage B2 tumors (P < .05) using one-way factorial analysis of variance and the Fisher test as a post-hoc comparison. No statistical difference was observed between primary tumors and paired synchronous colorectal liver metastasis (66% vs. 63%, respectively).
Frequency of AI in Each Microsatellite for Each Stage
To determine loci involved in the progression of the colorectal tumor, we analyzed the mean value of AI frequency for each microsatellite. The average frequency ranged from 26% to 81% (see Table 2). The alteration frequency of five microsatellites (D8S264, D9S179, TP53, D18S53, D18S61) exceeded 60%, in agreement with previous studies showing that losses of 18p, 18q, 8p, and p53 are the most frequent events observed in colorectal tumors.
In the groups of patients stratified by the Astler-Coller classification, three patterns of AI frequency evolution depending on stages could be distinguished. First, as expected, most of microsatellites (18/33) showed an increasing AI frequency correlated to tumor progression. These microsatellites were localized on 13 chromosomes 1q, 2q, 3p and 3q, 6p and 6q, 8p, 9p and 9q, 11q, 14q, 15q, 16q, 17q, 18q, and 22q. Among them, five markers (D1S305, D2S138, D3S1282, D17S790, D22S928) were significantly more altered in stage D than in stage A/B1 (P < .05). In addition, three markers (D8S264, D9S179, D18S61) of the five loci with an AI frequency of more than 60% in the overall population also showed an increase through stages, but the difference was not statistically significant. Second, 12 microsatellites showed AI frequencies roughly at the same level in all stages. These microsatellites were localized on chromosome arms 1p and 1q, 4p, 5p and 5q, 8q, 10p and 10 q, 13q, 17p and 17q, and 18p. Third, two microsatellites (D4S414, D20S107) showed a significant negative correlation in AI frequency between stage A/B1 and stage D (P = .033 and P = .044, respectively). One microsatellite (D1S207) showed a specific pattern in variation of AI frequency based on stage: in other words, the percentage of AI frequency decreased between stages A/B1 and B2, was stable between stages B2 and C, and strongly and significantly increased between stages C and D (P < .022). Further, the AI frequency observed in synchronous colorectal liver metastases, at several loci, was increased, with values reaching 100% for D18S61. The comparison between the 10 synchronous colorectal liver metastases and paired primary tumors did not reveal significant differences at the analyzed loci. Among these loci, D11S916 and D15S127 had been identified as significantly altered in a previous study comparing primary tumors with metachronous colorectal liver metastases. 27
DISCUSSION
In this study, we were able to show that the AI frequency increased in accord with Astler-Coller stages, but this increase was not linear. However, such analysis could represent a new, highly informative tool to classify colorectal tumors. Further, our systematic and sensitive approach allowed us to identify several highly altered loci up to 94%, such as D18S61 in stage D.
Identification of Patients With Low-Stage Tumors and Risk of Recurrence or Metastases
In a previous study, 27 using a smaller and different set of microsatellites, we identified three loci correlated with the tumor spread. In the current study, using more and for some of them different microsatellites, we focused on identifying a possible subpopulation of early tumors (stages A/B1-B2) with high AI frequency, mimicking the pattern observed in advanced stages. To make our results more comprehensive, we classified the patients depending on their AI frequency with a threshold of 35%, although no clear group could be distinguished except the group at 30% to 40%, which appeared slightly more represented. With our panel of 35 microsatellites, we were able to identify two populations of patients with tumors with a low or high level of alterations using this AI frequency threshold of 35%. Both types of tumors (low or high AI frequency) could be detected in every Astler-Coller stage. As expected, most of the highly altered tumors were significantly associated with stages C and D, confirming the relation between an accumulation of genetic alterations and tumor progression, as previously described. 7,10,11 Because it has been already described that recurrence or metastasis could develop in patients with stage A/B1 tumors, 31,32 new tools were required to identify patients with higher risks. Interestingly, with our panel of microsatellites, half (24/46) of the A/B1 and B2 tumors also showed a high level of AI frequency of more than 35%, whereas 15 of 46 tumors (7 A/B1, 8 B2) still had an AI frequency of more than 50%. Our molecular study allowed us to identify a group of patients with early tumors but high AI frequency. Using a digital SNP-PCR analysis, chromosome instabilities were already observed in colorectal adenomas, suggesting that multiple genomic instabilities can occur at very early stages; this confirms our observation that some “early” stages (A/B1) had a high frequency of alterations. 33 These patients could have a high potential of recurrence or metastatic evolution and would require more clinical investigations during follow-up, and eventually a specific regimen. However, in terms of the follow-up of A/B1 patients, this cohort is not yet sufficient to obtain statistically significant data. Further systematic multicenter studies are needed to establish correlations between survival and molecular status of the tumor to determine the importance of the molecular status as an independent prognosis factor.
Identification of New Markers for Prognosis
Our allelotyping allowed us to identify several loci with a clearly significant positive correlation with Astler-Coller stages. Among them, D1S305 is close to FGFR2 coding for the fibroblast growth receptor 2, which has been described as a transmembrane receptor implicated in tumor expansion, 34 and D2S138 is close to the WNT receptor, which has been described as regulating the APC pathway. 35,36 These loci appeared as good candidates for markers of tumor progression. Further clinical studies would confirm whether these markers could be considered as independent prognostic factors. Further, the analysis of AI frequency for each microsatellite has revealed that five loci (D8S264, D9S179, TP53, D18S53, D18S61) were altered in more than 60% of the overall population. Among them, three (D8S264, D9S179, D18S61) showed a progressive increase with tumor progression through stages. In terms of alteration at chromosome 18q, several studies have shown various percentages of alterations. Such discrepancies could be due either to different locations of the analyzed loci or to the preservation (frozen vs. paraffin-embedded formalin-fixed) of specimens. 37,38 Interestingly, D18S61 is close to the Bcl2 gene, suggesting an involvement of this gene in colorectal tumorigenesis; this is in agreement with the well-known antiapoptotic function of Bcl2 protein. 39–42 As for the locus D8S264, no target genes could be identified from the Genome Data Bank, and further studies are required to determine the role of this locus in tumor progression. However, chromosome arm 8p has already been shown to be involved in oncogenesis. 17,43 Between the two other highly rearranged loci, TP53 presented a roughly stable AI frequency through stages. The TP53 marker is informative for p53 protein because, localized in the first intron of the gene, the p53 gene has been described as a highly frequent and early-altered gene in the adenoma-to-carcinoma sequence. 44 We noticed that D5S346, which is informative for the APC gene, 45 showed, as expected, a high AI frequency in stages A/B1, with no further increase through tumor progression. The APC gene was shown to play an essential role in initiation of tumorigenesis during the hyperproliferative tissue to late adenoma sequence. 46–48 Our results suggest that this gene, involved in initiation, is not necessarily implicated in tumor progression.
Moreover, this study allowed us to describe a new pattern of the evolution of AI frequency through stages; in other words, a decrease of AI frequency during tumor progression. In fact, two loci (D4S414, D20S107) showed a negative correlation through stages. D20S107, significantly more altered in stage A/B1 versus B2 (P = .031), is close to the topoisomerase I gene, and topoisomerase I is mainly involved in transcription mechanisms. 49 This protein is a target of chemotherapy drugs, and identifying the gene status in colorectal tumors could help in determining chemotherapy regimens. 50
Further, concerning the AI frequency observed in synchronous liver metastases, several loci were highly altered. Although the increase was not significant for most of them, probably because of the low number of metastases, D11S916 and D15S127, for which the AI frequency doubled through stages to metastasis, were shown to be significantly altered in a previous study comparing primary with metachronous metastases. 27 Our results suggest that loci other than those, which showed an increased AI frequency through stages A to D, could be mainly involved in metastatic evolution. D11S916 is close to the TIMP-1 gene coding for an inhibitor of metalloproteinase, which has been shown to be involved in the control of extracellular matrix remodelling and cell migration. 51,52 Few studies have focused on the analysis of synchronous liver metastases and the corresponding primary colon carcinoma. 2,53,54 Additional studies characterizing alterations in both primary colorectal tumor and paired synchronous liver metastases would help us understand the mechanisms and functions involved in metastatic process. Together, these results allow us to characterize two patterns of evolution of AI frequency through stages, describing two types of function. A decrease in AI frequency at one microsatellite could indicate that alterations of genes located at this locus could have an effect mainly at the initiation of tumorigenesis, thus leading to a disadvantage of growth for tumor cell clones. Further studies would permit us to determine the favorable prognostic value of such alterations. Reciprocally, an increase in AI frequency, the most common situation, suggests a role of targeted genes at this locus in tumor invasion.
In conclusion, this study allows us to correlate an increase in AI to tumor progression. Moreover, a subgroup of patients with stage A/B1 tumors with a high level of genomic alterations was characterized. These patients could represent a high-risk group and could benefit from closer follow-up and more aggressive treatments. Routine molecular analyses are a powerful tool for the study of genomic features and could be used more and more in clinical and pathologic investigations to adapt therapeutic strategy. Finally, our study establishes the basis of a systematic strategy, allowing clarification of the use of such a molecular test as providing independent information about tumor classification and eventually risk factors.
Discussion
Prof. A. M. M. Eggermont: That was a beautiful presentation of an impressive study. Impressive because this is the era of molecular staging; you are right on target. This kind of study takes a lot of organization and logistics, infrastructure, contacts with your molecular biologists, department of pathology: I think it is a class act and I applaud you for doing such a large study.
To my knowledge, it is the first study that addresses the problem by looking at the LOH or allelic imbalance issue both in the primary tumors as well as on the metastatic sites, and you are the first one to come up with three markers which have an independent prognostic value apart from the one that was reported for the chromosome 18Q by Molly’s group in 1998.
You set a new stage, which indicates that there is going to be a lot of development in this field. Nevertheless, of course, there are also a number of questions that remain in the development of molecular staging. One of these questions is that LOH or allelic imbalance for breast cancer was to be a very potent independent prognostic factor for the 1p chromosome and this is a mutation or a loss that you also find in about 60% of the colorectal cancers. Did one of your 54 markers address this particular site, and do you have any additional information on that?
Second, sometimes a patient may have had a different primary tumor in the past and you may have been confronted with a single liver metastasis or with more than one liver tumor, and the question may have arisen, what is actually the origin of this particular liver tumor? I ask this because in my department, we do as a standard procedure for single masses in the lungs in patients who have in their past either had a head and neck tumor or breast cancer; for instance, you are facing the problem whether this is metastatic disease or a new primary.
We do LOH routinely on cytology of the mass in the lung and by LOH profiling we will know whether it is a metastasis or a primary tumor, so it is important for clinical decision making. Did you ever confront that problem in the context of this study?
Lastly, just a comment: are we just collecting stamps here in the start of this molecular staging development or, as your last slide seems to indicate, are we indeed discovering new markers on the basis of which we can make management decisions and treatment choices, and do we have new monitoring markers?
Thank you.
Dr. J. C. Weber (closing): Thank you for your kind comments and questions.
Among the 35 microsatellite markers, two targeted chromosome 1p, and we obtained some preliminary results. It is interesting to notice, for example, that concerning D1S207, we observed a special pattern in tumor samples. Indeed, we could show that there is a decrease in A1 frequency between stage A/B1 and B2, a stable level between stages B and C, and an increase between stages C and D, but only statistically significant between C and D stages. These results have only been shown for this microsatellite, and it is too early to draw definitive conclusions. Interestingly, in a previous study, topoisomerase I has been identified as a potential marker of prognosis in a population of 30 primary tumors without liver metastases. Topoisomerase I could be a prognostic marker. In case of a mutation, some regimens of chemotherapy could be nonefficient. We are now trying to correlate survival with our panel of markers, including topoisomerase I. This is still in progress.
Regarding the liver metastasis, there was no confusion possible with other types of metastases, as they were synchronous and resected (or a biopsy was taken) at the same time as the paired primary tumor. In addition, we are working now on synchronous liver metastases and paired primary tumors in order to better characterize molecular patterns and differences between both specimens.
In previous studies published by other authors, we noticed that most of the liver metastases analyzed were metachronous; so it is not sure that there is a correlation between primary tumor and liver tumor (nonpaired tumors). Moreover, chemotherapy had been sometimes applied between the resection of the primary tumor and the resection of the liver metastases, and this treatment could have induced modifications in the molecular pattern of colorectal liver metastases. For these reasons, one of the advantages of working on synchronous liver metastases is to collect the two samples (primary and metastases) at the same time.
For the new markers, we are trying to correlate survival with these markers, and we now have complete data for 93 patients. However, we would like to go further in order to confirm the significant correlation between molecular markers and survival in larger series.
Footnotes
Correspondence: Daniel Jaeck, MD, PhD, FRCS, Professor of Surgery, Centre de Chirurgie Viscérale et de Transplantation, Hôpitaux Universitaires de Strasbourg, Avenue Molière, 67098 Strasbourg Cedex, France.
E-mail: daniel.jaeck@chru-strasbourg.fr
Accepted for publication April 2001.
References
- 1.Vogelstein B, Fearon ER, Kern SE, et al. Allelotype of colorectal carcinomas. Science 1989; 244: 207–211. [DOI] [PubMed] [Google Scholar]
- 2.Ookawa K, Sakamoto M, Hirohashi S, et al. Concordant p53 and DCC alterations and allelic losses on chromosomes 13q and 14q associated with liver metastases of colorectal carcinoma. Int J Cancer 1993; 53: 382–387. [DOI] [PubMed] [Google Scholar]
- 3.Bardi G, Parada LA, Bomme L, et al. Cytogenetic comparisons of synchronous carcinomas and polyps in patients with colorectal cancer. Br J Cancer 1997; 76: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meijer GA, Hermsen MA, Baak JP, et al. Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridisation. J Clin Pathol 1998; 51: 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Angelis PM, Clausen OP, Schjolberg A, et al. Chromosomal gains and losses in primary colorectal carcinomas detected by CGH and their associations with tumour DNA ploidy, genotypes and phenotypes. Br J Cancer 1999; 80: 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100: 57–70. [DOI] [PubMed] [Google Scholar]
- 7.Fearon ER. Molecular genetic studies of the adenoma–carcinoma sequence. Adv Intern Med 1994; 39: 123–147. [PubMed] [Google Scholar]
- 8.Ding SF, Delhanty JD, Zografos G, et al. Chromosome allele loss in colorectal liver metastases and its association with clinical features. Br J Surg 1994; 81: 875–878. [DOI] [PubMed] [Google Scholar]
- 9.Di Renzo MF, Poulsom R, Olivero M, et al. Expression of the Met/hepatocyte growth factor receptor in human pancreatic cancer. Cancer Res 1995; 55: 1129–1138. [PubMed] [Google Scholar]
- 10.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996; 87: 159–170. [DOI] [PubMed] [Google Scholar]
- 11.Kochhar R, Halling KC, McDonnell S, et al. Allelic imbalance and microsatellite instability in resected Duke’s D colorectal cancer. Diagn Mol Pathol 1997; 6: 78–84. [DOI] [PubMed] [Google Scholar]
- 12.Nanashima A, Tagawa Y, Yasutake T, et al. Deletion of chromosome 11 and development of colorectal carcinoma. Cancer Detect Prev 1997; 21: 7–11. [PubMed] [Google Scholar]
- 13.Shigemori C, Wada H, Matsumoto K, et al. Tissue factor expression and metastatic potential of colorectal cancer. Thromb Haemost 1998; 80: 894–898. [PubMed] [Google Scholar]
- 14.Sparks AB, Morin PJ, Vogelstein B, et al. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res 1998; 58: 1130–1134. [PubMed] [Google Scholar]
- 15.Paredes-Zaglul A, Kang JJ, Essig YP, et al. Analysis of colorectal cancer by comparative genomic hybridization: evidence for induction of the metastatic phenotype by loss of tumor suppressor genes. Clin Cancer Res 1998; 4: 879–886. [PubMed] [Google Scholar]
- 16.Arai T, Akiyama Y, Yamamura A, et al. Allelotype analysis of early colorectal cancers with lymph node metastasis. Int J Cancer 1998; 79: 418–423. [DOI] [PubMed] [Google Scholar]
- 17.Halling KC, French AJ, McDonnell SK, et al. Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst 1999; 91: 1295–1303. [DOI] [PubMed] [Google Scholar]
- 18.Ionov Y, Peinado MA, Malkhosyan S, et al. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993; 363: 558–561. [DOI] [PubMed] [Google Scholar]
- 19.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science 1993; 260: 816–819. [DOI] [PubMed] [Google Scholar]
- 20.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for Cancer Detection and Familial Predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998; 58: 5248–5257. [PubMed] [Google Scholar]
- 21.Dietmaier W, Wallinger S, Bocker T, et al. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res 1997; 57: 4749–4756. [PubMed] [Google Scholar]
- 22.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 1999; 9: 89–96. [DOI] [PubMed] [Google Scholar]
- 23.Lukish JR, Muro K, DeNobile J, et al. Prognostic significance of DNA replication errors in young patients with colorectal cancer. Ann Surg 1998; 227: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messerini L, Ciantelli M, Baglioni S, et al. Prognostic significance of microsatellite instability in sporadic mucinous colorectal cancers. Hum Pathol 1999; 30: 629–634. [DOI] [PubMed] [Google Scholar]
- 25.Niederacher D, Picard F, van Roeyen C, et al. Patterns of allelic loss on chromosome 17 in sporadic breast carcinomas detected by fluorescent-labeled microsatellite analysis. Genes Chromosomes Cancer 1997; 18: 181–192. [DOI] [PubMed] [Google Scholar]
- 26.Schneider A, Borgnat S, Lang H, et al. Evaluation of microsatellite analysis in urine sediment for diagnosis of bladder cancer. Cancer Res 2000; 60: 4617–4622. [PubMed] [Google Scholar]
- 27.Schneider A, Rohr S, Kelly MD, et al. Microsatellite instability and allelic imbalance in primary and secondary colorectal cancer. Aust NZ J Surg 2000; 70: 587–592. [DOI] [PubMed] [Google Scholar]
- 28.Sugano K, Nakashima Y, Yamaguchi K, et al. Sensitive detection of loss of heterozygosity in the TP53 gene in pancreatic adenocarcinoma by fluorescence-based single-strand conformation polymorphism analysis using blunt-end DNA fragments. Genes Chromosomes Cancer 1996; 15: 157–164. [DOI] [PubMed] [Google Scholar]
- 29.Delattre O, Olschwang S, Law DJ, et al. Multiple genetic alterations in distal and proximal colorectal cancer. Lancet 1989; 2: 353–356. [DOI] [PubMed] [Google Scholar]
- 30.Olschwang S, Hamelin R, Laurent-Puig P, et al. Alternative genetic pathways in colorectal carcinogenesis. Proc Natl Acad Sci USA 1997; 94: 12122–12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michelassi F, Block GE, Vannucci L, et al. A 5- to 21-year follow-up and analysis of 250 patients with rectal adenocarcinoma. Ann Surg 1988; 208: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson RM, Perencevich NP, Malcolm AW, et al. Patterns of recurrence following curative resection of adenocarcinoma of the colon and rectum. Cancer 1980; 45: 2969–2674. [DOI] [PubMed] [Google Scholar]
- 33.Shih IM, Zhou W, Goodman SN, et al. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res 2001; 61: 818–822. [PubMed] [Google Scholar]
- 34.Jayson GC, Vives C, Paraskeva C, et al. Coordinated modulation of the fibroblast growth factor dual receptor mechanism during transformation from human colon adenoma to carcinoma. Int J Cancer 1999; 82: 298–304. [DOI] [PubMed] [Google Scholar]
- 35.Polakis P. Wnt signaling and cancer. Genes Dev 2000; 14: 1837–1851. [PubMed] [Google Scholar]
- 36.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell 2000; 103: 311–320. [DOI] [PubMed] [Google Scholar]
- 37.Carethers JM, Hawn MT, Greenson JK, et al. Prognostic significance of allelic loss at chromosome 18q21 for stage II colorectal cancer. Gastroenterology 1998; 114: 1188–1195. [DOI] [PubMed] [Google Scholar]
- 38.Naidoo R, Tarin M, Chetty R. A comparative microsatellite analysis of colorectal cancer in patients <35 years and >50 years of age. Am J Gastroenterol 2000; 95: 3266–3275. [DOI] [PubMed] [Google Scholar]
- 39.Giarnieri E, Nagar C, Valli C, et al. BCL2 and BAX expression in hyperplastic and dysplastic rectal polyps. Hepato-Gastroenterology 2000; 47: 159–162. [PubMed] [Google Scholar]
- 40.Kroemer G. The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat Med 1997; 3: 614–620. [DOI] [PubMed] [Google Scholar]
- 41.Pereira H, Silva S, Juliao R, et al. Prognostic markers for colorectal cancer: expression of P53 and BCL2. World J Surg 1997; 21: 210–213. [DOI] [PubMed] [Google Scholar]
- 42.Ishijima N, Miki C, Ishida T, et al. The immunohistochemical expression of BCL-2 oncoprotein in colorectal adenocarcinoma. Surg Today 1999; 29: 682–684. [DOI] [PubMed] [Google Scholar]
- 43.Lerebours F, Olschwang S, Thuille B, et al. Deletion mapping of the tumor suppressor locus involved in colorectal cancer on chromosome band 8p21. Genes Chromosomes Cancer 1999; 25: 147–153. [DOI] [PubMed] [Google Scholar]
- 44.Soussi T. The p53 tumor suppressor gene: from molecular biology to clinical investigation. Ann NY Acad Sci 2000; 910: 121–139. [DOI] [PubMed] [Google Scholar]
- 45.Spirio L, Joslyn G, Nelson L, et al. A CA repeat 30–70 KB downstream from the adenomatous polyposis coli (APC) gene. Nucleic Acids Res 1991; 19: 6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith KJ, Johnson KA, Bryan TM, et al. The APC gene product in normal and tumor cells. Proc Natl Acad Sci USA 1993; 90: 2846–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spirio L, Nelson L, Ward K, et al. A CA-repeat polymorphism close to the adenomatous polyposis coli (APC) gene offers improved diagnostic testing for familial APC. Am J Hum Genet 1993; 52: 286–296. [PMC free article] [PubMed] [Google Scholar]
- 48.Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta 1997; 1332: F127–147. [DOI] [PubMed] [Google Scholar]
- 49.Wang JC. DNA topoisomerases. Annu Rev Biochem 1985; 54: 665–697. [DOI] [PubMed] [Google Scholar]
- 50.Arbuck SG, Takimoto CH. An overview of topoisomerase I-targeting agents. Semin Hematol 1998; 35: 3–12. [PubMed] [Google Scholar]
- 51.Gomez DE, Alonso DF, Yoshiji H, et al. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur J Cell Biol 1997; 74: 111–122. [PubMed] [Google Scholar]
- 52.Gomis-Ruth FX, Maskos K, Betz M, et al. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature 1997; 389: 77–81. [DOI] [PubMed] [Google Scholar]
- 53.Thorstensen L, Qvist H, Nesland JM, et al. Allelotype profiles of local recurrences and distant metastases from colorectal-cancer patients. Int J Cancer 1996; 69: 452–456. [DOI] [PubMed] [Google Scholar]
- 54.Blaker H, Graf M, Rieker RJ, et al. Comparison of losses of heterozygosity and replication errors in primary colorectal carcinomas and corresponding liver metastases. J Pathol 1999; 188: 258–262. [DOI] [PubMed] [Google Scholar]