Abstract
We report the cloning and sequence analysis of Cerastes vipera and Macrovipera lebetina transmediterranea cDNAs coding for short non-RGD (Arg-Gly-Asp) disintegrins and for dimeric disintegrin subunits. The mRNAs belong to the short-coding class, suggesting that these disintegrin mRNAs may be more widely distributed than previously thought. Our data also argue for a common ancestry of the mRNAs of short disintegrins and those coding for precursors of dimeric disintegrin chains. The Macrovipera lebetina transmediterranea dimeric disintegrin reported to inhibit the laminin-binding integrins α3β1, α6β1 and α7β1 was analysed using a proteomic approach and was shown to bear MLD (Met-Leu-Asp) and VGD (Val-Gly-Asp) motifs. The results highlight the fact that disintegrins have evolved a restricted panel of integrin-blocking sequences that segregate with defined branches of the phylogenetic tree of the integrin α-chains, providing novel insights into the evolutionary adaptation of the snake venom antagonists to the ligand-binding sites of their target integrin receptors.
Keywords: Cerastes vipera, Macrovipera lebetina transmediterranea, integrin inhibition, snake venom disintegrin, snake venomics, toxin evolution
Abbreviations: a.m.u., atomic mass units; CID, collision-induced dissociation; CV-n, cDNA clone n coding for a Cerastes vipera disintegrin; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; ML-n, cDNA clone n coding for a Macrovipera lebetina transmediterranea disintegrin; MS/MS, tandem MS; SVMP, snake venom Zn2+-metalloproteinase; TFA, trifluoroacetic acid; VLO5, disintegrin from the venom of Macrovipera lebetina obtusa (=Vipera lebetina obtusa)
INTRODUCTION
Snake venom glands are very specialized tissues that possess a high capacity of protein secretion, and are a rich source of pharmacologically active proteins [1]. According to their major toxic effect in an envenomed animal, snake venoms may be conveniently classified as neurotoxic and haemorrhagic. Among the first group are the Elapidae snakes (mambas, cobras and, particularly, the Australian snakes, which are well known to be the most toxic in the world). On the other hand, venoms of Viperidae and Crotalidae snakes (vipers and rattlesnakes) contain a number of pro- and anti-coagulant proteins. Procoagulant proteins, such as activators of prothrombin, plasminogen, Factors V, IX and X, and fibrinolytic enzymes, activate the coagulation system, causing the consumption of coagulation factors. Anticoagulant protein can be grouped into a few major protein families, including enzymes (serine proteinases, Zn2+-dependent metalloproteinases of the reprolysin family, L-amino acid oxidase and group II phospholipase A2 isoenzymes) and proteins with no enzymatic activity (C-type lectins, protease inhibitors and disintegrins) [2,3]. Depending on the venom composition, the pro- and the anti-coagulant proteins, working synergistically with other toxins [e.g. myotoxins, growth factors, CRISP (cysteine-rich secretory protein), bradykinin-potentiating peptides], cause a diversity of clinical effects, including local tissue swelling and necrosis, flaccid paralysis, systemic myolysis, cardiotoxicity, renal failure and different coagulopathies (i.e. persistent bleeding, thrombosis, thrombocytopenia) [4].
SVMPs (snake venom Zn2+-metalloproteinases) are multi-domain proteins which diverged from the extracellular region of membrane-associated ADAMs (a disintegrin and metalloproteinases) and were recruited into the venom proteome at the base of the Colubroidea radiation 60–80 million years ago [5–7]. Disintegrins, a family of small (40–100 amino acids) cysteine-rich polypeptides that selectively block the adhesive function of integrin receptors [8,9], are released in the venoms of various vipers by proteolytic processing of PII SVMP precursors [10,11] or are synthesized from short-coding mRNAs [12]. Currently, disintegrins can be classified according to their length and number of disulphide bonds [13]. The first group includes short disintegrins composed of 41–51 residues and four disulphide bonds. The second group is formed by the medium-sized disintegrins which contain approx. 70 amino acids and six cystine bonds. The third group includes long disintegrins with an ∼84-residue polypeptide cross-linked by seven disulphide bridges. The fourth group is composed of homo- and hetero-dimers. Dimeric disintegrins contain subunits of approx. 67 residues with ten cysteines involved in the formation of four intrachain disulphides and two interchain cystine linkages [14,15]. Like many other venom toxins, the integrin-inhibitory activity of disintegrins depends on the appropriate pairing of cysteine residues which determines the conformation of the inhibitory loop that harbours an active tripeptide located at the apex of a mobile loop protruding 14–17 Å (1 Å=0.1 nm) from the protein core [8,9]. The current view is that the structural diversity of disintegrins has been achieved during ophidian evolution through gene duplications followed by mutations and deletions of gene regions coding for cysteine residues that are engaged in the formation of disulphide bonds in the ancestor molecules [13].
The reports of two different mRNA structures (PII precursor and a short mRNA) coding for dimeric disintegrin subunits prompted us to search for mRNAs encoding Cerastes vipera and Macrovipera lebetina transmediterranea [in the present paper, the EMBL reptile database (http://www.reptile-database.org/) snake species nomenclature is employed] disintegrins [16]. Our results show that short-coding mRNAs may be more widely distributed than previously thought, perhaps representing the canonical structure of dimeric and short disintegrin precursors. This information, in turn, is relevant for our understanding of the genomic basis of the molecular mechanism underlying the structural diversification of disintegrins and their adaptation to the ligand-binding architecture of their target integrin receptors.
MATERIALS AND METHODS
cDNA cloning and sequencing
Total RNA was extracted from pooled venom glands of C. vipera and M. lebetina transmediterranea. The vipers were captured in the Sahara desert in the south of Tunisia (C. vipera) and in the rocky mountains of the north of Tunisia (M. lebetina transmediterranea) and were kept in captivity at the Serpentarium of the Institut Pasteur de Tunis. Snakes were killed 3 days after venom extraction, when toxin gene transcription rates are at a peak. Glands were homogenized under liquid nitrogen and total RNA was extracted using guanidinium thiocyanate/phenol/chloroform as described in [17]. The first strand of cDNA was reverse-transcribed using a standard method and the following reaction mixture: 12 μl of 0.1% DEPC (diethylpyrocarbonate)-treated water containing 0.1 μg of 5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTT16-3′ (Qt) (purchased from Sigma-Genosys) and 1 μg of total venom RNA was heated to 70 °C for 10 min to denature any possible secondary RNA structure, cooled to ice temperature, and mixed with 4 μl of 5× First Strand Buffer (Promega), 2 μl of 0.1 M dithiothreitol (Promega), 1 μl of RNAsin (ribonuclease inhibitor) (40 units/μl) (Promega), 1 μl of dNTP (10 mM each) (Eppendorf) and 200 units of M-MLV RT (Moloney murine leukaemia virus reverse transcriptase) (RNase H−; Promega). The final volume was adjusted to 20 μl, and the reaction mixture was incubated at 42 °C for 1 h, followed by 10 min at 50 °C to inactivate the enzyme. Disintegrin-coding DNAs were amplified by PCR using venom cDNAs as template and the following pairs of primers. (i) Forward primer, 5′-CGTGCCATGGATTGTACAACTGGACCATG-3′, which contains the sequence coding for the first six residues of the short disintegrin jerdostatin (Swiss-Prot/TrEMBL accession number AY262730) and the CCATGG NcoI restriction site, and reverse primer, 5′-GCCTCGAGTATTAGCCATTCCCGGGATAAC-3′, which includes a stop codon (in bold italics), the CTCGAG restriction site for XhoI, and the last six C-terminal residues of jerdostatin. (ii) Forward primer, 5′-ATGATCCA(A/G)GTTCTCTTGG-3′ (synthesized according to the highly conserved pro-peptide of metalloproteinase precursors), and reverse primer, 5′-GAGGACTCGAGCTCAAGC-3′ (Qt-anchor). (iii) Forward primer, 5′- CGTGCCATGGATAATTCTGCACATCCATG-3′, which includes the NcoI restriction site and the nucleotide sequence coding for the N-terminal amino acid sequence (NSAHPC) of the dimeric disintegrin CV-3 [16], and reverse primer, 5′-GCCTCGAGTATTAGCCATTCCCGGGATAAC-3′, as above.
The PCR protocol included an initial denaturation step at 95 °C for 2 min followed by 35 cycles of denaturation (10 s at 94 °C), annealing (20 s at 50 °C) and extension (30 s at 72 °C), and a final extension for 10 min at 72 °C. The amplified fragments were separated by 2% agarose gel electrophoresis and purified using the Perfect Pre Gel Clean Up kit (Eppendorf) and were cloned in either a pGEM-T vector (Promega) or a pET32a vector (Novagen), which were used to transform Escherichia coli DH5α cells (Novagen) by electroporation using an Eppendorf 2510 electroporator following the manufacturer's instructions. Positive clones, selected by growing the transformed cells in LB (Luria–Bertani) broth containing 10 μg/ml ampicillin, were confirmed by PCR-amplification using the above primers, and the sequences of the inserts were subjected to sequencing on an Applied Biosystems model 377 DNA sequencing system.
Isolation and characterization of venom proteins
Venoms of C. vipera and M. lebetina transmediterranea were collected from snakes of both sexes kept in captivity at the Serpentarium of the Institut Pasteur de Tunis. After extraction, the venoms were immediately freeze-dried and stored at 4 °C. For reverse-phase HPLC separations, 2–5 mg of the crude venom was dissolved in 100 μl of 0.05% TFA (trifluoroacetic acid) and 5% acetonitrile, and insoluble material was removed by centrifugation in an Eppendorf centrifuge at 13000 g for 10 min at room temperature (25 °C). Proteins in the soluble material were separated using an ETTAN™ LC HPLC system (Amersham Biosciences) and a Lichrospher RP100 C18 column (250 mm×4 mm, 5 μm particle size) eluted at 1 ml/min with a linear gradient of TFA in water (solution A) and acetonitrile (solution B) using the following chromatographic conditions: isocratically (5% solution B) for 20 min, followed by 5–55% solution B for 68 min, and 55–70% solution B for 20 min. Protein detection was at 215 nm, and peaks were collected manually and dried in a SpeedVac (Savant). The purity and molecular mass of the reverse-phase-isolated proteins were checked by SDS/13% PAGE, N-terminal sequencing (using an Applied Biosystems Procise 492 sequencer), MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS using a Voyager-DE Pro instrument (Applied Biosystems) and 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid) (Sigma) saturated in 70% acetonitrile and 0.1% TFA as matrix, and electrospray ionization MS with a triple quadrupole-ion trap hybrid instrument (QTrap from Applied Biosystems) equipped with a nanospray source (Protana).
In-gel enzymatic digestion and mass fingerprinting
SDS/13% PAGE-separated Coomassie Brilliant Blue-stained protein bands were subjected to automated digestion with sequencing-grade bovine pancreas trypsin (Roche) at a final concentration of 20 ng/μl in 50 mM ammonium bicarbonate, pH 8.3, using a ProGest digestor (Genomic Solutions) following the manufacturer's instructions. Digestions were done before reduction with dithiothreitol (10 mM for 15 min at 65 °C) and carbamidomethylation with iodoacetamide (50 mM for 60 min at room temperature). The tryptic peptide mixtures were dried in a SpeedVac, the samples were dissolved in 5 μl of 50% acetonitrile and 0.1% TFA, and were subjected to mass fingerprinting. When necessary, the digestion mixtures were diluted with 0.1% TFA to a final acetonitrile concentration of <10% and were freed from reagents using a C18 Zip-Tip pipette tip (Millipore), following the manufacturer's instructions.
For mass fingerprinting analysis, 0.85 μl of the digests were spotted on to a MALDI–TOF sample holder, mixed with an equal volume of a saturated solution of α-cyano-4-hydroxycinnamic acid (Sigma) in 70% acetonitrile containing 0.1% TFA, dried, and analysed with an Applied Biosystems Voyager-DE Pro MALDI–TOF mass spectrometer, operated in delayed extraction and reflector modes. Database searches were constrained to a mass tolerance of 100 p.p.m. A tryptic peptide mixture of Cratylia floribunda seed lectin (Swiss-Prot accession number P81517) prepared and characterized previously in our laboratory was used as mass-calibration standard (mass range 450–3300 Da).
CID (collision-induced dissociation) by MS/MS (tandem MS)
For peptide sequencing, the protein digest mixture was subjected to electrospray ionization MS/MS analysis using a QTrap mass spectrometer [18] equipped with a nanospray source. Doubly charged ions selected after Enhanced Resolution MS analysis were fragmented using the Enhanced Product Ion with Q0 trapping option at 250 a.m.u. (atomic mass units)/s across the entire mass range. For MS/MS experiments, Q1 was operated at unit resolution, the Q1–Q2 collision energy was set to 35 eV, the Q3 entry barrier was 8 V, the LIT (linear ion trap) Q3 fill time was 250 ms, and the scan rate in Q3 was 1000 a.m.u./s. CID spectra were interpreted manually or using the online form of the MASCOT program at http://www.matrixscience.com.
RESULTS AND DISCUSSION
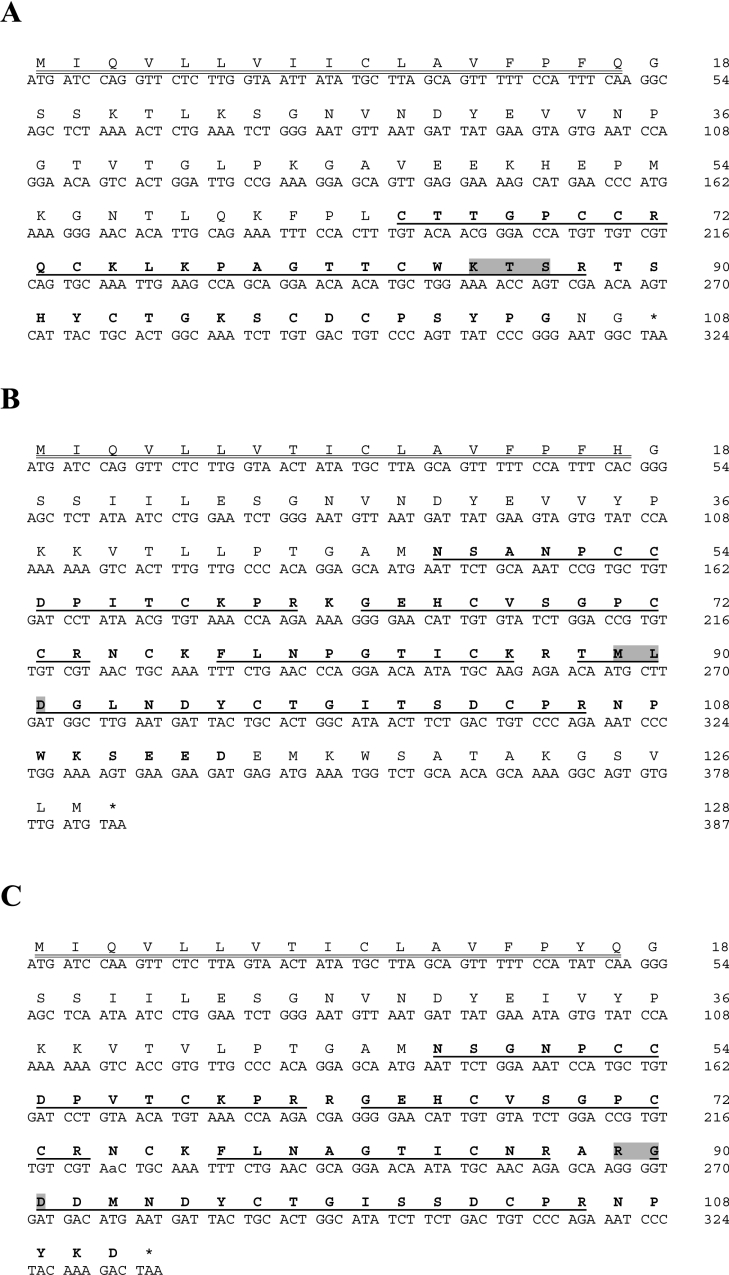
Full-length disintegrin mRNAs were cloned from cDNA libraries constructed from C. vipera (CV) and M. lebetina transmediterranea (ML) venom gland polyadenylated mRNA. The deduced amino acid sequences of the CV and ML cDNAs are displayed in Figures 1 and 2 respectively. The DNA sequences of clones CV-short, CV-11, CV-1, ML-(6,9,10), ML-(2,8,15) and ML-3 are accessible from the Swiss-Prot/TrEMBL database (http://us.expasy.org) under accession numbers AM114012, AM114013, AM114014, AM114015, AM114016 and AM114017 respectively.
Figure 1. DNA and deduced amino acid sequences of C. vipera disintegrins.
The nucleotide sequences are numbered in the 5′→3′ direction from the initial codon ATG to the stop codon TGA. The predicted mature protein sequences are in boldface. The integrin-binding tripeptide sequences are shaded. *, stop; (A) Clone CV-short amplified using the primer combination (i) and a pGEM-T vector. (B) Full-length sequence of clone CV-11 amplified using primers (ii) in a pGEM-T vector. The signal sequence is double-underlined and the deduced disintegrin sequence is in boldface. (C) Clone CV-1, amplified with primers (iii) using a pET32a cloning vector. The deduced disintegrin sequence is in boldface. The upstream sequence correspond to thioredoxin and to the polyhistidine sequence (in italics) of the expression vector.
Figure 2. Full-length DNA and deduced amino acid sequences of M. lebetina transmediterranea disintegrins.
The nucleotide sequences, amplified using primers (ii) in a pGEM-T vector, are numbered in the 5′→3′ direction from the initial codon ATG to the stop codon TGA. The predicted mature protein sequences are in boldface. The signal sequences are double-underlined. (A) Clones ML-(6,9,10); the experimentally determined N-terminal sequence is underlined. (B) Clones ML-(2,8,15). (C) Clone ML-3. Amino acid sequences identified in tryptic peptides of dimeric disintegrins ML-9/10 and in ML-11/12 isolated from M. lebetina transmediterranea venom (Table 1) are underlined in (B) and (C) respectively.
cDNAs encoding short non-RGD (Arg-Gly-Asp) disintegrins
cDNAs coding for short non-RGD-containing disintegrins were amplified from both venom gland libraries (Figures 1A and 2A). The 132 nucleotides of the CV-short clone (Figure 1A) (Swiss-Prot/TrEMBL accession number AM114012) encoding a short RTS (Arg-Thr-Ser)-disintegrin are 100% identical with that of jerdostatin from Trimeresurus jerdonii (Swiss-Prot/TrEMBL accession number AY262730) (Figure 3). This short disintegrin has not been detected when the C. vipera venom composition was analysed using a proteomic approach [16], and may thus represent a non-expressed mRNA. On the other hand, the ML-(6,9,10) (Swiss-Prot/TrEMBL accession number AM114015) cDNA-deduced amino acid sequence 1–41 (Figure 3) (calculated monoisotopic M+H+: 4408.1 Da) corresponds to the venom protein ML-4 [16], a KTS (Lys-Thr-Ser)-short disintegrin whose monoisotopic mass determined by MALDI–TOF MS was 4408.8 Da. This protein has been also called lebestatin by Kallech-Ziri et al. [19] and represents approx. 1% of the total venom proteins [16].
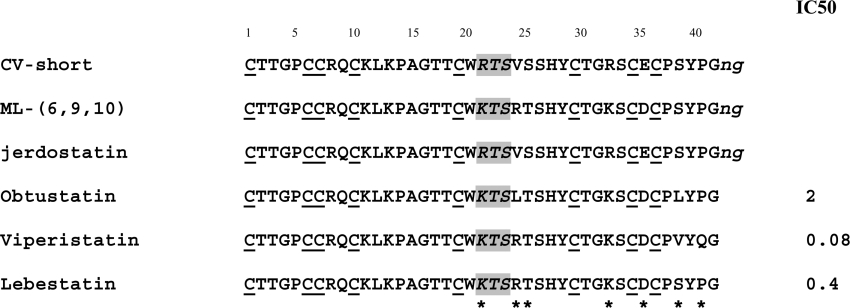
Figure 3. Alignment of the sequences of short KTS/RTS-disintegrins.
Comparison of the amino acid sequences of the short disintegrins deduced from the cDNA clones CV-short (C. vipera) and ML-(6,9,10) (M. lebetina transmediterranea) with jerdostatin (T. jerdonii), obtustatin (M. lebetina obtusa), viperistatin (V. palestinae) and lebestatin (M. lebetina transmediterranea). Cysteine residues are underlined, and the integrin-inhibitory motifs are in italics and shaded. The C-terminal dipeptide of the cDNA sequences, which are processed in the mature disintegrins obtustatin, viperistatin and lebestatin, are shown in italics and in lower case. The IC50 values (in nM) of inhibition of the adhesion of α1-transfected cells to immobilized collagen IV are shown for obtustatin [21], lebestatin [19] and viperistatin [22].
The CV-short and ML-(6,9,10) encoded disintegrins display high amino acid sequence similarity to the KTS-disintegrins obtustatin, from Macrovipera lebetina obtusa venom [20,21], and viperistatin, isolated from the venom of Vipera palestinae [22] (Figure 3). Noteworthily, all of the seven amino acid residues which depart from the sequences of CV-short, ML-(6,9,10), obtustatin and viperistatin are segregated within the C-terminal half of the molecule, including the integrin-recognition loop and the C-terminal tail, two structural elements which form a conformational functional epitope in the three-dimensional structure of obtustatin [23,24]. In particular, the CV-short disintegrin contains an RTS motif in the place of the KTS tripeptide found in obtustatin, ML-(6,9,10) (lebestatin) and viperistatin. The KTS motif has been shown to endow disintegrins with selective inhibitory activity of the in vitro adhesion of integrin α1β1 to immobilized collagen IV [19,21,22] and of angiogenesis in vivo [19,20]. Recombinant native-folded jerdostatin also exhibits restricted inhibitory selectivity towards integrin α1β1, and its RTS motif appears to represent a 4-fold more potent inhibitory motif than KTS [25]. Disintegrins viperistatin and lebestatin express within their integrin-binding loops an RTS sequence C-terminal of the KTS tripeptide. Synthetic peptides representing the entire integrin-binding loops of obtustatin (CWKTSLTSHYC) and viperistatin (CWKTSRTSHYC) showed IC50 values of inhibition of the adhesion of α1-transfected K562 cells to immobilized collagen IV of 101 and 645 μM respectively [22]. Whether viperistatin and lebestatin use the KTS or the RTS motif to impair the ligand-binding ability of α1β1 remains puzzling. However, a comparison of their amino acid sequences and inhibitory activities (Figure 3) strongly suggest that the enhanced potency of viperistatin over lebestatin may be due to their different amino acid residues (Val38/Ser and Gln40/Pro) within the C-terminal tails.
cDNAs encoding dimeric disintegrin chains
The cDNA-deduced amino acid sequences from clones CV-11 (Swiss-Prot/TrEMBL accession number AM114013) and CV-1 (Swiss-Prot/TrEMBL accession number AM114014) (Figures 1B and 1C), and ML-(2,8,15) (Swiss-Prot/TrEMBL accession number AM114016) and ML-3 (Swiss-Prot/TrEMBL accession number AM114017) (Figures 2B and 2C), show large similarity to known sequences of subunits of dimeric disintegrins, including the number and position of the ten cysteine residues [13] (Figure 4). Disintegrins are produced from larger mosaic PII SVMP precursors [10,11] or are synthesized from short-coding mRNAs [12]. Our results showing that all full-length C. vipera and M. lebetina transmediterranea dimeric disintegrin precursors are encoded by short mRNAs indicate that short-coding mRNAs may be more widely distributed than previously thought, perhaps representing the canonical structure of dimeric disintegrin precursors. In support of this view, the two dimeric disintegrin mRNAs found in the transcriptome of Bitis gabonica, gabonin-1 and gabonin-2 [26], and all the full-length cDNAs coding for dimeric disintegrin subunits characterized from a venom gland cDNA library of Echis ocellatus (P. Juárez, S. C. Wagstaff, L. Sanz, R. A. Harrison and J. J. Calvete, unpublished work), belong to the short-coding region class. Our working hypothesis is that removal of the metalloproteinase domain may have represented a step in the evolutionary pathway yielding dimeric disintegrins from a PII-metalloproteinase-coding precursor.
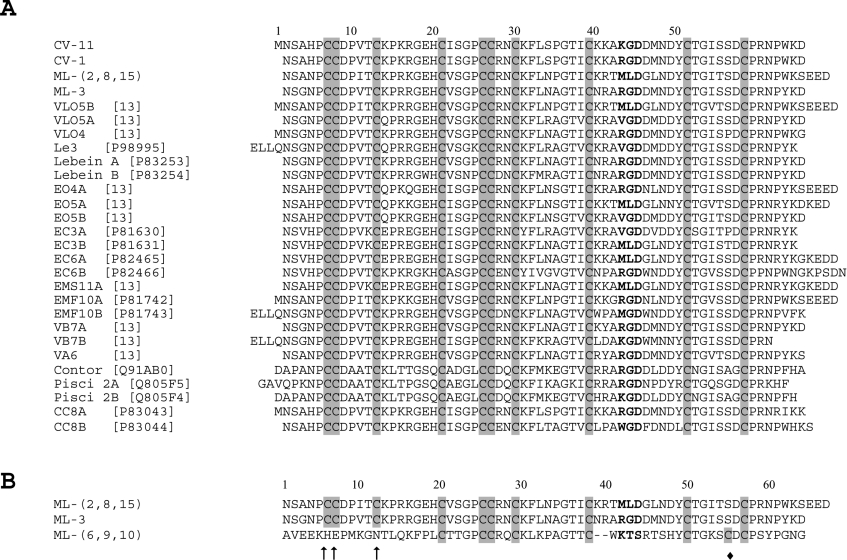
Figure 4. Multiple sequence analysis of disintegrins.
(A) Multiple amino acid sequence alignment of the C. vipera (CV-) and M. lebetina transmediterranea (ML-) cDNA-deduced disintegrin sequences with those of selected dimeric disintegrin subunits. Cysteine residues are shadowed in grey. The integrin-binding tripeptides are shown in boldface. References and, when available, the Swiss-Prot/TrEMBL (http://us.expasy.org/sprot/) database accession numbers are given in square brackets. (B) Alignment of the cDNA-deduced amino acid sequences of the ML-(2,8,15) and the ML-3 cDNA-deduced dimeric disintegrin subunit sequences and the short KTS-containing disintegrin precursor encoded in clone ML-(6,9,10). Amino acid positions which in dimeric disintegrin subunits correspond to conserved cysteine residues are marked with an arrow. The short-disintegrin-specific cysteine residue is labelled ◆.
Clones CV-11 and CV-1 encode dimeric disintegrin subunits departing in just the first amino acid of their integrin-inhibitory motifs, RGD in CV-11 and KGD in CV-1 (Figure 4A). A single reverse-phase HPLC peak (CV-3) containing dimeric disintegrin(s) of molecular mass 14 kDa has been reported in the venom proteome of C. vipera [16]. To establish the subunit combinations of these C. vipera venom disintegrins, the isolated CV-3 peak was submitted to electrospray ionization MS. Major ions corresponded to proteins of molecular masses (±1 Da) of 14027.2, 14157.1 and 14318.1 Da. This clearly indicates the presence of both heterodimers (N2SAH…RGD…WKD65/N2SAH…KGD…WKD65; calculated molecular mass 14026 Da; M1NSAH…RGD…WKD65/N2SAH…KGD…WKD65 and/or N2SAH…RGD…WKD65/M1NSAH…KGD…WKD65; calculated molecular mass 14157 Da) and a homodimer (M1NSAH…RGD…WKD65/M1NSAH…RGD…WKD65; calculated molecular mass 14316 Da).
M. lebetina transmediterranea venom has been reported to contain dimeric disintegrins [16,27,28]. Clone ML-3 (Swiss-Prot/TrEMBL accession number AM114017) displayed in Figure 2(C) codes for the α-chain of lebein (Swiss-Prot accession number P83253) [27]. The MLD (Met-Leu-Asp)-disintegrin encoded in clone ML-(2,8,15) (Figure 2B) departs in only three residues from the B-chain of VLO5 from M. lebetina obtusa [13] (Figure 4). The other M. lebetina transmediterranea cDNA clones reported here code for novel dimeric disintegrin subunits. To investigate their presence in the dimeric disintegrins expressed in the venom, the venom proteins were fractionated by reverse-phase HPLC and the dimeric disintegrins (peaks ML-9, ML-10, ML-11 and ML-12 in [16]) were submitted to N-terminal sequencing, molecular-mass determination using electrospray ionization MS and to structural characterization by MALDI–TOF mass fingerprinting followed by CID MS/MS of tryptic peptide ions. Peaks ML-9 (major disintegrin peak in ML venom; 14080.8±0.5 Da) and ML-10 (minor peak; 13711.0±2 Da), both had the N-terminal sequence (NSGNPCCDPVTCK…) and tryptic peptide maps expected for the heterodimeric disintegrin lebein [27], also called MS-II by Eble et al. [28] (Table 1). Peaks ML-11 (14617.2±0.8 Da) and ML-12 (14748.4±2 Da) were also shown to correspond to homologous proteins by mass fingerprinting and exhibited an identical N-terminal sequence (NSANPCCDPITCKPRKGEHCVSGPCCRNCKFLNPGTICK…). MS/MS analysis of ML-11- and ML-12-derived tryptic peptides (Table 1) indicated that these disintegrins were heterodimers of the MLD-containing subunit encoded by clone ML-(2,8,15) (Figure 2B) and an N-terminally blocked VGD (Val-Gly-Asp)-bearing subunit structurally related to the disintegrin domain of the fibrinolytic proteinase lebetase-3 from M. lebetina transmediterranea [29], which in turn is identical with the A-chain of VLO5 [13]. Thus M. lebetina transmediterranea ML-12 may be highly homologous with VLO5 from M. lebetina obtusa (MALDI–TOF mass 14728 Da) [13]. The mass difference of approx. 130 Da between ML-11 and ML-12 may correspond to C-terminal (i.e. ±glutamate) processing.
Table 1. Amino acid sequence determination by CID MS/MS of peptide ions generated by tryptic digestion of reduced and carbamidomethylated dimeric disintegrins isolated from M. lebetina transmediterranea venom.
*, The dimeric disintegrins are termed according to the nomenclature of Bazaa et al. [16]. Z, pyrrolidone carboxylic acid; m, methionine sulphoxide.
| Disintegrin (HPLC peak)* | m/z of tryptic ion (z) | Amino acid sequence | Protein identified |
|---|---|---|---|
| ML-9, ML-10 | 881.9 (2+) | NSGNPCCDPVTCKPR | Lebein(α=β)-(1–15) |
| 660.3 (2+) | GEHCVSGPCCR | Lebeinα-(17–27) | |
| 583.8 (2+) | FLNAGTICNR | Lebeinα-(31–40) | |
| 982.4 (2+) | GDDMNDYCTGISSDCPR | Lebein(α=β)-(43–59) | |
| 897.8 (2+) | GWHCVSNPCCDNCK | Lebeinβ-(17–30) | |
| ML-11, ML-12 | 525.9 (2+) | FLNPGTICK | Clone ML-(2,8,15) |
| 660.4 (2+) | GEHCVSGPCCR | Clone ML-(2,8,15) | |
| 896.1 (2+) | NSANPCCDPITCKPR | Clone ML-(2,8,15) | |
| 730.7 (3+) | TMLDGLNDYCTGITSDCPR | Clone ML-(2,8,15) | |
| 634.3 (3+) | ZNSANPCCDPITCQPR | No match | |
| 717.5 (3+) | AVGDDmDDYCTGISSDCPR | Lebetase-3 (VLO5A) |
ML-11 and ML-12 display the N-terminal sequence of the β-subunit of lebein-2 (or MS-I; 14735 Da) [28]. In addition, and like ML-11/12, the α-subunit of lebein-2 had a blocked N-terminus [28]. The mass difference between lebein-2 and ML-12 may be due to oxidation of a methionine residue as determined in the tryptic peptide ion 717.5 (3+) (Table 1). Interestingly, lebein-2 impaired the binding of integrins α3β1, α6β1 and α7β1 to their cognate laminin isoform ligands with IC50 values of 39–220 nM, and the attachment of myoblasts to laminin and to fibronectin [28]. Eble et al. [28] also noticed that a 44 kDa protein, possibly corresponding to lebetase precursor, interacted in far-Western blot assays with the α7β1 integrin. As a whole, these data indicate that an MLD motif harbours the blocking activity of lebein-2 (ML-11/12) towards integrins α3β1, α6β1 and α7β1, while the VGD motif may target the fibronectin receptor of the α5β1 integrin.
A pathway for the evolution of lebestatin
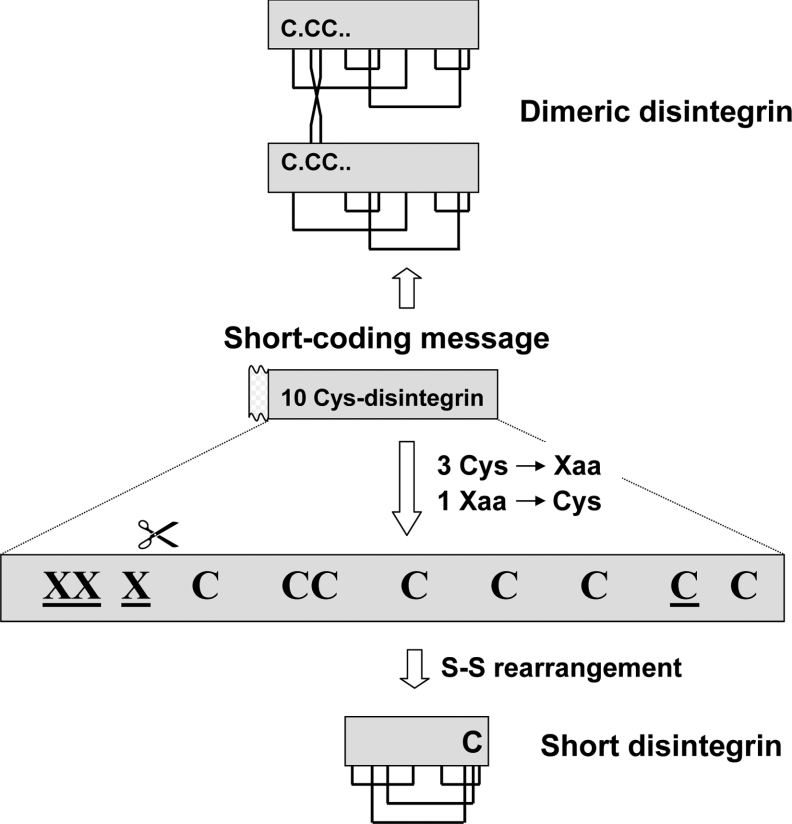
In a previous study, we performed a phylogenetic analysis of the disintegrin family and have proposed that the different disintegrin subfamilies have evolved through a disulphide bond engineering mechanism (see Figure 4 in [13]). However, the lack of full-length cDNA sequences of short disintegrin precursors impeded us in distinguishing whether this class of disintegrins arose from long, medium-sized or dimeric subunits (see Figure 5 in [13]). The realization that the short KTS-disintegrin lebestatin (clone ML-6,9,10), the jerdostatin precursor (Swiss-Prot/TrEMBL accession number AY262730), and possibly the RTS-disintegrin (CV-short clone) too, are coded for by short-coding precursors strongly argues for a common ancestry of the mRNAs of these short disintegrins and those coding for precursors of dimeric disintegrin chains. Comparison of the cDNA-deduced sequences of the lebestatin precursor with those of dimeric disintegrin subunits (Figure 4B) indicates that the first three cysteine residues of the latter have been replaced in the short-disintegrin precursor by His6, Glu7 and Asn12. These mutations hinder formation of the two interchain cystine linkages, which in schistatin (Echis carinatus sochureki) correspond to Cys7A-Cys12B and Cys12A-Cys7B [15,30] and in EMF-10 (Eristocophis macmahoni) are Cys7A-Cys7B and Cys12A-Cys12B [14]. A further substitution of cysteine for the residue located two positions N-terminal of the last cysteine residue of the dimeric disintegrin subunit [residue 55 of ML-(6,9,10) and ML-3; Figure 4B], followed by proteolytic processing of the N- and the C-terminal regions, are further molecular events needed for generating the mature short-disintegrin lebestatin (Figure 5). In both cDNAs, ML-(6,9,10) and ML-3, residue 55 is a serine coded for by a TCT codon. Serine is also the most common residue in the position that is topologically equivalent to the short-disintegrin-specific cysteine residue (56 in the multiple alignment shown in Figure 4(A) and marked with a black rhombus in Figure 4B), and it is worthy of notice that substitution of cysteine (TGC or TGT) for serine (TCC or TCT) can be accomplished by a single G→C mutation.
Figure 5. An evolutionary pathway for lebestatin.
Proposed common ancestry of the mRNA precursors coding for the short disintegrin lebestatin and dimeric disintegrin subunits. The proposed evolutionary pathway includes replacement of three N-terminal cysteine residues by other amino acids, along with the appearance of a cysteine residue between the ninth and the tenth cysteine of the precursor, proteolytic processing of the N-terminal region and the formation of short-disintegrin-specific disulphide bonds.
Disintegrins have evolved a restricted panel of integrin-inhibitory motifs
Snakes have developed an efficient arsenal of integrin receptor antagonists. Thus, with the exception of the α2β1 integrin, which is targeted by a number of C-type lectin-like proteins [31], inhibitory motifs towards β1 and β3 integrins have evolved in different members of the disintegrin family [8,9]. Among them, RGD blocks the β1 and β3 integrins α8β1, α5β1, αvβ1, αvβ3 and αIIbβ3; MLD targets the α4β1, α4β7 and α9β1 integrins; VGD and MGD impair the function of the α5β1 integrin; KGD inhibits the αIIbβ3 integrin with a high degree of selectivity; WGD has been reported to be a potent inhibitor of the RGD-dependent integrins α5β1, αvβ3 and αIIbβ3; and KTS and RTS represent selective α1β1 inhibitors. The realization that lebein-2 (MLD and VGD motifs) antagonizes the activity of the α3β1, α6β1, α7β1 and α5β1 integrins reveals an almost complete repertoire of integrin β1- and β3-binding motifs (Figure 6).
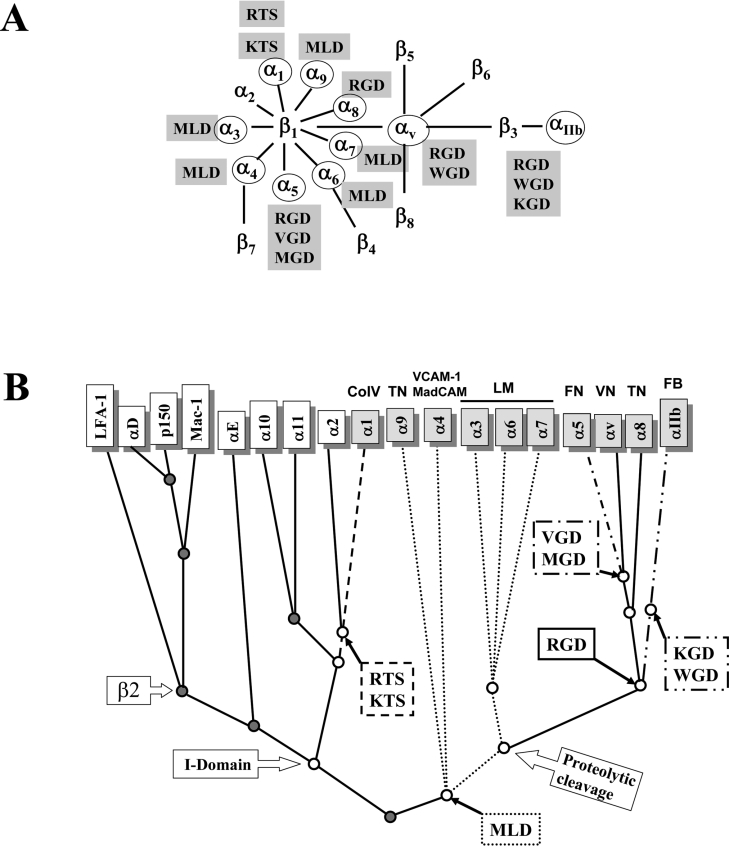
Figure 6. Proposed evolutionary adaptation of integrin-inhibitory motifs to the ligand-binding sites of integrins.
(A) Diagram of the integrin family and the different disintegrin tripeptide motifs that block specific integrin–ligand interactions. Integrin heterodimers antagonized by snake venom disintegrins are encircled. Integrins are a major class of cell-surface transmembrane type I receptors that mediate a wide variety of cell–cell and cell–extracellular matrix interactions [34]. The integrin α and the β subunits combine in a restricted manner to form different dimers, each of which exhibits a distinct ligand-binding profile. α1β1 is a receptor for collagen IV (CoIV); α2β1 binds collagen I; α4β1 interacts with fibronectin (FN) and vascular cell adhesion molecule 1 (VCAM-1); α4β7 bind the same ligands as α4β1 and in addition is a receptor for mucosal addressin cell adhesion molecule (MadCAM); α5β1 represents the major fibronectin receptor; integrins α3β1, α6β1, and α7β1 represent major laminin (LM) receptors; α8β1 and α9β1 bind tenascin (TN); αvβ1 and αvβ3 are major vitronectin (VN) receptors; and αIIbβ3 is the platelet fibrinogen (FB) receptor that is involved in platelet aggregation. (B) Segregation of disintegrin inhibitory motifs within the phylogenetic tree of the integrin α-subunits are highlighted by identical box and branch line patterns. Integrin receptors which are targets of disintegrins are in grey boxes and their major ligand are indicated. Integrin-inhibitory sequence motifs and their integrin receptor targets are linked by thick solid arrows. The phylogenetic relationships have been adapted from data in [35,36]. Branches are not scaled according to evolutionary distance. Branch points linked with the emergence of the proteolytic cleavage of integrin α-subunits, and the acquisition of an αI-domain (I-domain), are indicated.
The crystal structure of the extracellular segment of integrin αvβ3 in complex with an RGD ligand [32] showed that the peptide fits into a crevice between the αV propeller and the β3 A-domain. The arginine side chain is held in place by interactions with αV carboxylates 218 and 150, the glycine residue makes several hydrophobic interactions with αV, and the aspartate ligand interacts primarily with βA residues. Thus, with the exception of the KTS and RTS α1β1 inhibitory motifs, the conserved aspartate residue might be responsible for the binding of disintegrins to integrin receptors which share a β subunit, while the two other residues of the integrin-binding motif (RG, KG, MG, WG, ML or VG) may dictate the integrin specificity through interaction with the α-subunit. Figure 6(A) shows a diagram of the integrin family and the reported disintegrin motifs that block specific integrin–ligand interactions. In addition, Figure 6(B) shows the clear segregation of the different integrin-inhibitory motifs with defined branches of the phylogenetic tree of the integrin α-chains. This highlights the fact that disintegrins have evolved an efficient, albeit restricted, panel of integrin blocking sequences, and provides novel insights into the evolutionary adaptation of the snake venom antagonists to the ligand-binding sites of integrin receptors.
The cluster of the αI-domain-containing integrins segregates into two monophyletic groups, including the collagen receptors α1, α2, α10 and α11, and the leucocyte-specific integrins αX, αL, αM, αD and αE (Figure 6B). The αI-domain-containing integrins that mediate direct adhesion to collagens seem to have a relatively recent history, since they have been found only in the vertebrate clade [33]. Our proposed model forecasts that the (K/R)TS-disintegrins may represent the most recent group of the disintegrin family and that members of this group may also act as antagonists of the αI-domain-containing collagen-binding integrins α10β1 and α11β1.
Acknowledgments
This study has been partly financed by grant BFU2004-01432/BMC from the Ministerio de Educación y Ciencia, Madrid, Spain, and by the Spanish–Tunisian Cooperation Programme 29p/02 from the Agencia Española de Cooperación Internacional (AECI).
References
- 1.Markland F. S. Snake venoms and the hemostatic system. Toxicon. 1998;36:1749–1800. doi: 10.1016/s0041-0101(98)00126-3. [DOI] [PubMed] [Google Scholar]
- 2.Ménez A. Chichester, UK: John Wiley & Sons Ltd; 2002. Perspectives in Molecular Toxinology. [Google Scholar]
- 3.Juárez P., Sanz L., Calvete J. J. Snake venomics: characterization of protein families in Sistrurus barbouri venom by cysteine mapping, N-terminal sequencing, and tandem mass spectrometry analysis. Proteomics. 2004;4:327–338. doi: 10.1002/pmic.200300628. [DOI] [PubMed] [Google Scholar]
- 4.White J. Snake venom and coagulopathy. Toxicon. 2005;45:951–967. doi: 10.1016/j.toxicon.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 5.Moura-da-Silva A. M., Theakston R. D. G., Crampton J. M. Evolution of disintegrin cysteine-rich and mammalian matrix-degrading metalloproteinases: gene duplication and divergence of a common ancestor rather than convergent evolution. J. Mol. Evol. 1996;43:263–269. doi: 10.1007/BF02338834. [DOI] [PubMed] [Google Scholar]
- 6.Fry B. G., Wüster W. Assembling an arsenal: origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol. Biol. Evol. 2004;21:870–883. doi: 10.1093/molbev/msh091. [DOI] [PubMed] [Google Scholar]
- 7.Fry B. G. From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15:403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvete J. J., Marcinkiewicz C., Monleón D., Esteve V., Celda B., Juárez P., Sanz L. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Calvete J. J. Structure–function correlations of snake venom disintegrins. Curr. Pharm. Des. 2005;11:829–835. doi: 10.2174/1381612053381783. [DOI] [PubMed] [Google Scholar]
- 10.Kini R., Evans H. J. Structural domains in venom proteins: evidence that metalloproteinases and nonenzymatic platelet aggregation inhibitors (disintegrins) from snake venoms are derived by proteolysis from a common precursor. Toxicon. 1992;30:265–293. doi: 10.1016/0041-0101(92)90869-7. [DOI] [PubMed] [Google Scholar]
- 11.Shimokawa K., Jai L.-G., Wang X.-M., Fox J. W. Expression, activation and sequencing of the recombinant snake venom metalloproteinase, pro-atrolysin E. Arch. Biochem. Biophys. 1996;335:283–294. doi: 10.1006/abbi.1996.0509. [DOI] [PubMed] [Google Scholar]
- 12.Okuda D., Koike H., Morita T. A new gene structure of the disintegrin family: a subunit of dimeric disintegrin has a short coding region. Biochemistry. 2002;41:14248–14254. doi: 10.1021/bi025876s. [DOI] [PubMed] [Google Scholar]
- 13.Calvete J. J., Moreno-Murciano M. P., Theakston R. D. G., Kisiel D. G., Marcinkiewicz C. Snake venom disintegrins: novel dimeric disintegrins and structural diversification by disulphide bond engineering. Biochem. J. 2003;372:725–734. doi: 10.1042/BJ20021739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calvete J. J., Jürgens M., Marcinkiewicz C., Romero A., Schrader M., Niewiarowski S. Disulfide bond pattern and molecular modelling of the dimeric disintegrin EMF-10, a potent and selective integrin α5β1 antagonist from Eristocophis macmahoni venom. Biochem. J. 2000;345:573–581. [PMC free article] [PubMed] [Google Scholar]
- 15.Bilgrami S., Tomar S., Yadav S., Kaur P., Kumar J., Jabeen T., Sharma S., Singh T. P. Crystal structure of schistatin, a disintegrin homodimer from saw-scaled viper (Echis carinatus) at 2.5 Å resolution. J. Mol. Biol. 2004;341:829–837. doi: 10.1016/j.jmb.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 16.Bazaa A., Marrakchi N., El Ayeb M., Sanz L., Calvete J. J. Snake venomics: comparative analysis of the venom proteomes of the Tunisian snakes Cerastes cerastes, Cerastes vipera and Macrovipera lebetina. Proteomics. 2005;5:4223–4235. doi: 10.1002/pmic.200402024. [DOI] [PubMed] [Google Scholar]
- 17.Harrison R. A., Oliver J., Hasson S. S., Bharati K., Theakston R. D. G. Novel sequences encoding venom C-type lectins are conserved in phylogenetically and geographically distinct Echis and Bitis viper species. Gene. 2003;315:95–102. doi: 10.1016/s0378-1119(03)00716-9. [DOI] [PubMed] [Google Scholar]
- 18.Le Blanc J. C., Hager J. W., Ilisiu A. M., Hunter C., Zhong F., Chu I. Unique scanning capabilities of a new hybrid linear ion trap mass spectrometer (QTRAP) used for high sensitivity proteomics applications. Proteomics. 2003;3:859–869. doi: 10.1002/pmic.200300415. [DOI] [PubMed] [Google Scholar]
- 19.Kallech-Ziri O., Luis J., Daoud S., Bazaa A., Srairi N., Andreotti N., Lehman M., Zouari R., Mabrouk K., Marvaldi J., et al. Lebestatin, a short disintegrin from Macrovipera lebetina venom, inhibits integrin-mediated adhesion, migration of tumor cells and in vivo angiogenesis. Lab. Invest. 2005;85:1507–1516. [Google Scholar]
- 20.Marcinkiewicz C., Wainreb P. H., Calvete J. J., Kisiel D. G., Mousa S. A., Tuszynski G. P., Lobb R. R. Obtustatin: a potent selective inhibitor of α1β1 integrin in vitro and angiogenesis in vivo. Cancer Res. 2003;63:2020–2023. [PubMed] [Google Scholar]
- 21.Moreno-Murciano P., Monleón D., Calvete J. J., Celda B., Marcinkiewicz C. Amino acid sequence and homology modeling of obtustatin, a novel non-RGD-containing short disintegrin isolated from the venom of Vipera lebetina obtusa. Protein Sci. 2003;12:366–371. doi: 10.1110/ps.0230203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kisiel D. G., Calvete J. J., Katzhendel J., Fertala A., Lazarovici P., Marcinkiewicz C. Structural determinants of the selectivity of KTS-disintegrins for the α1β1 integrin. FEBS Lett. 2004;577:478–482. doi: 10.1016/j.febslet.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 23.Moreno-Murciano M. P., Monleón D., Marcinkiewicz C., Calvete J. J., Celda B. NMR solution structure of the non-RGD disintegrin obtustatin. J. Mol. Biol. 2003;329:135–145. doi: 10.1016/s0022-2836(03)00371-1. [DOI] [PubMed] [Google Scholar]
- 24.Monleón D., Moreno-Murciano M. P., Kovacs H., Marcinkiewicz C., Calvete J. J., Celda B. Concerted motions of the integrin-binding loop and the C-terminal tail of the non-RGD disintegrin obtustatin. J. Biol. Chem. 2003;278:45570–45576. doi: 10.1074/jbc.M307030200. [DOI] [PubMed] [Google Scholar]
- 25.Sanz L., Chen R.-Q., Pérez A., Hilario R., Juárez P., Marcinkiewicz C., Monleón D., Celda B., Xiong Y.-L., Pérez-Payá E., Calvete J. J. cDNA cloning and functional expression of jerdostatin, a novel RTS-disintegrin from Trimeresurus jerdonii and a specific antagonist of the α1β1 integrin. J. Biol. Chem. 2005;280:40714–40722. doi: 10.1074/jbc.M509738200. [DOI] [PubMed] [Google Scholar]
- 26.Francischetti I. M. B., My-Pham V., Harrison J., Garfield M. K., Ribeiro J. M. C. Bitis gabonica (Gaboon viper) snake venom gland: toward a catalog for the full-length transcripts (cDNA) and proteins. Gene. 2004;337:55–69. doi: 10.1016/j.gene.2004.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasmi A., Srairi N., Guermazi S., Dkhil H., Karoui H., El Ayeb M. Amino acid structure and characterization of a heterodimeric disintegrin from Vipera lebetina venom. Biochim. Biophys. Acta. 2001;1547:51–56. doi: 10.1016/s0167-4838(01)00168-6. [DOI] [PubMed] [Google Scholar]
- 28.Eble J. A., Bruckner P., Mayer U. Vipera lebetina venom contains two disintegrins inhibiting laminin-binding β1 integrins. J. Biol. Chem. 2003;278:26488–26496. doi: 10.1074/jbc.M301860200. [DOI] [PubMed] [Google Scholar]
- 29.Siigur E., Aaspollu A., Tu A. T., Siigur J. cDNA cloning and deduced amino acid sequence of fibrinolytic enzyme (lebetase) from Vipera lebetina snake venom. Biochem. Biophys. Res. Commun. 1996;224:229–236. doi: 10.1006/bbrc.1996.1012. [DOI] [PubMed] [Google Scholar]
- 30.Bilgrami S., Yadav S., Sharma S., Perbandt M., Betzel C., Singh T. P. Crystal structure of the disintegrin heterodimer from saw-scaled viper (Echis carinatus) at 1.9 Å resolution. Biochemistry. 2005;44:11058–11066. doi: 10.1021/bi050849y. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa T., Chijiwa T., Oda-Ueda N., Ohno M. Molecular diversity and accelerated evolution of C-type lectin-like proteins from snake venom. Toxicon. 2005;45:1–14. doi: 10.1016/j.toxicon.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 32.Xiong J.-P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. The crystal structure of the extracellular segment of integrin αvβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 33.Hughes A. L. Evolution of the integrin α and β families. J. Mol. Evol. 2001;52:63–72. doi: 10.1007/s002390010134. [DOI] [PubMed] [Google Scholar]
- 34.Hynes R. O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 35.Ewan R., Huxley-Jones J., Mould A. P., Humphries M. J., Robertson D. L., Boot-Handford R. P. The integrins of the urochordate Ciona intestinalis provide novel insights into the molecular evolution of the vertebrate integrin family. BMC Evol. Biol. 2005;5:31. doi: 10.1186/1471-2148-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huhtala M., Heino J., Casciari D., de Luise A., Johnson M. S. Integrin evolution: insights from ascidian and teleosts fish genomes. Matrix Biol. 2005;24:83–95. doi: 10.1016/j.matbio.2005.01.003. [DOI] [PubMed] [Google Scholar]