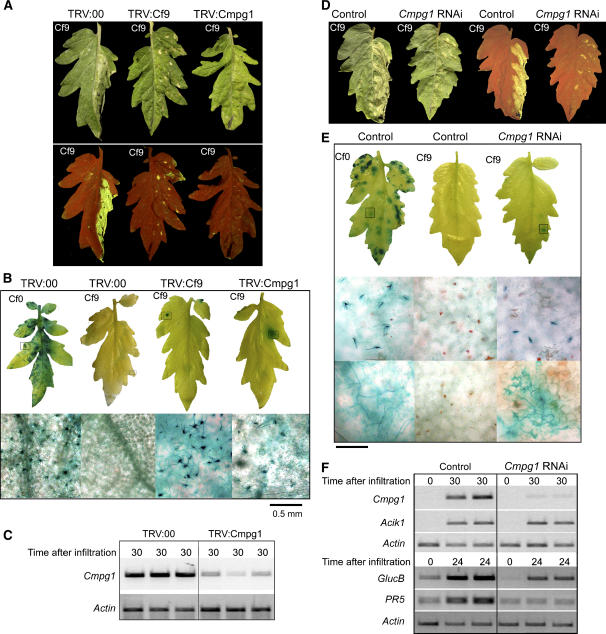
Figure 7.
Tomato Cmpg1 Is Required for Full Cf-9–Mediated Resistance to C. fulvum Expressing Avr9.
(A) VIGS in tomato. Transgenic Cf9 tomato cotyledons were inoculated with agrobacteria carrying the indicated TRV construct. Three weeks after infiltration, the right half of the tomato leaf was infiltrated with IF(+Avr9), and pictures were taken 3 d after infiltration under daylight (top panel) or UV light (bottom panel).
(B) Transgenic Cf9 or Cf0 tomato seedlings were silenced with the indicated TRV construct as described in (A). Three weeks later, plants were infected with C. fulvum race 4 GUS. Leaves were stained with X-gluc 3 weeks after C. fulvum inoculation, when pictures were taken (top panel). The fungal cycle was complete, as indicated by the conidiophores extruding from the stomata observed under the microscope (bottom panel).
(C) Tomato Cmpg1 transcript levels were analyzed by RT-PCR. Leaf discs from the indicated silenced tomato plants were harvested 30 min after infiltration with water. Total RNA was extracted and used for RT-PCR analysis with Sl Cmpg1-specific primers. Equal amounts of cDNA were used, as shown by the amplification with the constitutively expressed Actin gene.
(D) RNAi in tomato. Cf9 tomato plants were transformed with pHellsgate containing a tomato Cmpg1 hairpin. One leaf half from the transgenic (Cmpg1 RNAi) and untransformed (control) Cf9 tomato plants was infiltrated with IF(+Avr9) for HR induction. Pictures were taken 3 d after infiltration under daylight (left leaves) and UV light (right leaves).
(E) Cf0 and Cf9 untransformed (control) and Cf9 transgenic (Cmpg1 RNAi) tomato plants were infected with C. fulvum race 4 GUS. Leaves were stained with X-gluc 3 weeks after infection (top panel). Fungal cycle was complete, as indicated by the conidiophores extruding from the stomata observed under the microscope (middle panel). Hyphal growth inside the leaves was confirmed under the microscope (bottom panel).
(F) Tomato Cmpg1 mRNA levels were analyzed by RT-PCR. Leaf discs from the indicated tomato lines were harvested 30 min after IF(+Avr9) infiltration and processed as described in (C). Additionally, equal induction treatment is shown by the amplification of Sl Acik1. From the same leaves, samples were harvested after 24 h, and transcript levels for the defense-related genes GlucB and PR5 were determined using gene-specific primers.