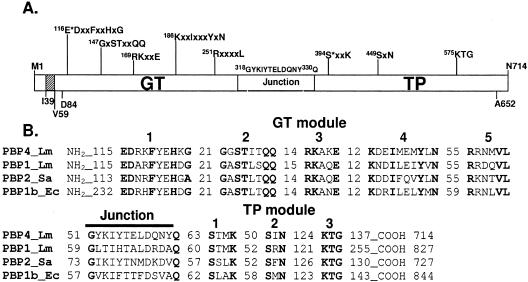
FIG. 1.
Modular organization of PBP4 and comparison of its amino acid sequence with those of other class A PBPs. (A) The catalytic GT and TP modules are separated by the linker motif (G318 to Q330). The conserved motifs of each module are shown. E116 is the proposed catalytic residue of the GT domain, and S394 is the active serine of the TP domain. The protein contains a short cytosolic tail (M1 to 38) followed by one predicted membrane anchor segment (I39 to V59). (B) Amino acid sequence alignment of L. monocytogenes PBP4 (Q8Y547) with L. monocytogenes (Lm) PBP1 (Q8Y610), S. aureus (Sa) PBP2 (Q53724), and E. coli (Ec) PBP1b (P02919). The conserved motifs and the number of amino acids between them or at the protein end are shown.