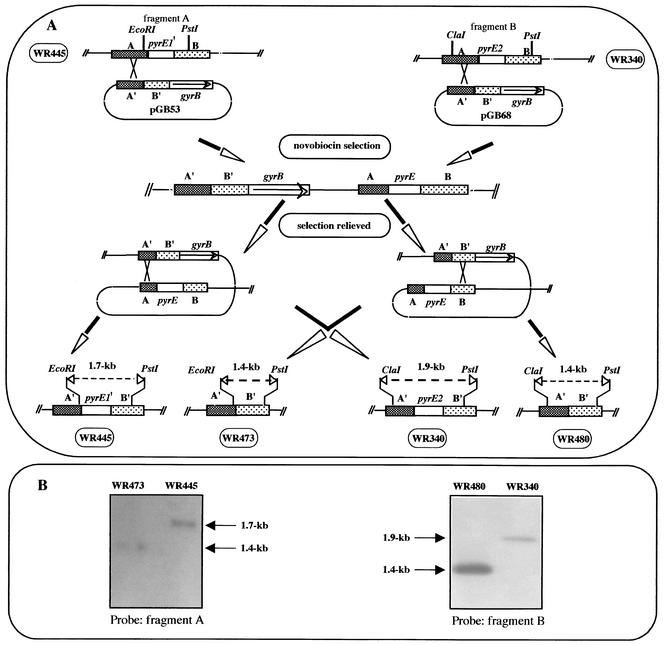
FIG. 3.
Schematic diagram of disruption of the pyrE1 gene in strain WR445 and disruption of the pyrE2 gene in strain WR340 and Southern blot analyses of the mutant strains. (A) Plasmids pGB53 and pGB68 were constructed as described in Materials and Methods. The plasmids were integrated into the chromosome by homologous recombination between the chromosomal sequence A and the plasmid sequence A′, and novobiocin-resistant recombinants were selected. Following relief of selection, recombination events could result in either reconstitution of the wild-type allele or deletion of the chromosomal pyrE gene. pyrE1′ is the first 330 bp of the pyrE1 gene. (B) Analysis of pyrE1 mutant. Total DNA was prepared from WR445 (parental strain) and WR473 (ΔpyrE1), digested with the EcoRI and PstI restriction enzymes, and analyzed by Southern blotting by using fragment A as a probe. For analysis of the ΔpyrE2 mutant, total DNA was prepared from WR430 (parental strain) and WR480 (ΔpyrE2), digested with the ClaI and PstI restriction enzymes, and analyzed by Southern blotting by using fragment B as a probe.