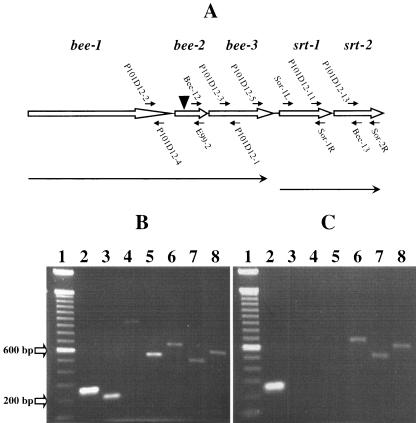
FIG. 2.
RT-PCR analyses to assess the expression and transcriptional linkage of genes within the bee locus in the wild-type strain E99 and mutant P101D12. (A) Schematic of the bee locus. The bee locus is comprised of three genes, bee-1, bee-2, and bee-3, encoding putative cell wall-anchored proteins, downstream of which are two ORFs, srt-1 and srt-2, encoding putative sortase enzymes. The locations of various primers used for the RT-PCR analysis are indicated. The position of the Tn917 insertion (▾) in mutant P101D12 is also indicated. Line arrows indicate putative transcriptional units. (B) Total RNA extracted from E99 was reverse transcribed using random hexamers and reverse transcriptase. An aliquot of the cDNA was then amplified by PCR with gene-specific primers. Primers P101D12-2/P101D12-4 (lane 2), P101D12-3/P101D12-1 (lane 3), P101D12-2/E99-2 (lane 4), Bee-12/P101D12-1 (lane 5), Sor-1L/Sor-1R (lane 6), P101D12-13/Sor-2R (lane 7), and P101D12-11/Bee-13 (lane 8) yielded 259-, 203-, 954-, 532-, 650-, 485-, and 581-bp amplification products, respectively. This implies that the genes bee-1, bee-2, and bee-3 may be cotranscribed as one transcriptional unit, with srt-1 and srt-2 cotranscribed independently. (C) Total RNA extracted from mutant strain P101D12 was used for RT-PCR. Primers P101D12-2/P101D12-4 (lane 2) yielded an expected 259-bp amplification product, indicating that bee-1 was being transcribed. However, no amplification products were obtained when primer pairs P101D12-3/P101D12-1 (lane 3), P101D12-2/E99-2 (lane 4), or Bee-12/P101D12-1 (lane 5) were used for amplification, suggesting that Tn917 insertion had abrogated not only the expression of bee-2 but also that of the downstream gene bee-3. Sor-1L/Sor-1R (lane 6), P101D12-13/Sor-2R (lane 7), and P101D12-11/Bee-13 (lane 8) yielded 650-, 485-, and 581-bp amplification products, respectively, implying that the Tn917 insertion in bee-2 did not abrogate expression of srt-1 and srt-2. For size reference, a 100-bp DNA ladder (Invitrogen, Carlsbad, CA) was used (lane 1).