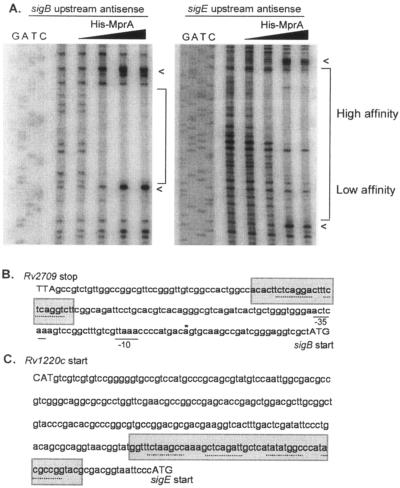
FIG. 3.
DNase I footprints of sigB and sigE upstream regions. (A) 32P-labeled DNA from the sigB and sigE upstream regions was incubated in the absence or presence of 1.3 to 260 pmol His-MprA∼P and treated with 0.2 units DNase I; arrowheads denote sites of DNase I hypersensitivity. (B) Protected regions (shaded areas) are depicted for both sigB (B) and sigE (C) upstream regions along with the known transcriptional start site (*) and −10 and −35 sites (solid lines). Putative 8-bp direct repeat motifs (dotted line) were determined by GCG analyses and were similar to those found upstream of mprA and pepD. Start and stop codons are depicted in capital letters.