Abstract
Microarray-based comparisons of three Mycobacterium avium subsp. paratuberculosis isolates, including one sheep strain and two cattle strains, identified three large genomic deletions in the sheep strain, totaling 29,208 bp and involving 24 open reading frames. These deletions may help explain some of the differences in pathogenicity and host specificity observed between the cattle and sheep strains of Mycobacterium avium subsp. paratuberculosis.
Mycobacterium avium subsp. paratuberculosis is the causative agent of Johne's disease and is known to exist as two phenotypically different strains, designated the sheep (S) and cattle (C) strains (2). Although a number of genomic-scale differences have recently been identified between these strains (1, 2, 3, 7, 10, 11), the specific genes involved and the mechanisms of host specialization of these strains have remained unclear.
The aim of the present study was to comprehensively identify and define the genomic differences between the S and C strains of M. avium subsp. paratuberculosis using a whole-genome M. avium subsp. paratuberculosis microarray. Results from these studies have uncovered two novel large-sequence polymorphisms as well as confirmed a deletion previously identified in the S strain by representational difference analysis (5).
Microarray analyses were undertaken to compare one S strain (Telford 9.2) and two C strains (CM00/416 and 316v) of M. avium subsp. paratuberculosis (Table S1 in the supplemental material) with DNA from the M. avium subsp. paratuberculosis K10 cattle isolate (4). M. avium subsp. paratuberculosis isolates were cultured and prepared for genomic extraction as previously described (5). All DNA samples were digested using the restriction endonuclease Sau3AI (5). DNA samples from the three M. avium subsp. paratuberculosis isolates were then compared with the K10 DNA in Cy3 and Cy5 dye swap hybridizations using hybridization conditions based on those described previously (6). Arrays were scanned using an arrayWoRxe optical scanner (Applied Precision), and the data were analyzed using softWoRx Tracker image analysis software. Further analysis was achieved by exporting the raw TIFF files of the scanned arrays into the softWoRx Tracker program. After normalization, genes corresponding to array locations that showed no hybridization with sheep genomic DNA were labeled as absent from that sheep strain. Finally, open reading frames (ORFs) that were not identified in either Cy3 or Cy5 dye swap hybridizations or not represented by at least two spot replicates were censored and not included in further analysis.
No differences were observed between the K10 strain and the CM00/416 and 316v C strains; however, 20 ORFs were found to be absent from the S strain (Telford 9.2). Among these 20 ORFs were two isolated ORFs (MAP0456 and MAP2325) and two clusters of ORFs spanning the regions from MAP1484c to MAP1488c and from MAP1728c to MAP1743c. PCR analysis was undertaken to examine the 20 ORFs within these putative deletions and the ORFs flanking them, using primers specific for each ORF (primers 1 to 74) (Table S2 in the supplemental material) and genomic DNA from the S (Telford 9.2) and C (CM00/416) strains as a template. Repeated PCR amplification experiments showed that MAP0456 was present in both the C and S strains, contradicting the microarray result for this ORF. The presence of ORFs MAP1734 and MAP1742c in the C strain could not be confirmed as neither ORF amplified, which may have been the result of using suboptimal PCR conditions for these loci. In all other instances, results of PCR amplification concurred with the microarray hybridization data, confirming the absence of MAP2325 and the MAP1484c-MAP1488c region in the S strain. Amplification experiments further extended the MAP1484c-MAP1488c region by two additional complete ORFs (MAP1489c and MAP1490) and a partial ORF (MAP1491). Similarly, PCR amplification confirmed the absence of the region extending from MAP1728c to MAP1743c in the S strain and included one additional ORF (MAP1744). The deletions in the S strain are referred to as deletion 1 (MAP1484c-MAP1491), deletion 2 (MAP1728c-MAP1744), and deletion 3 (MAP2325) and are described in detail in Table 1.
TABLE 1.
Summary of S strain deletions with reference to the M. avian subsp. paratuberculosis K10 genome
| S strain deletion | Start position | End position | Size (bp) | ORP(s) includeda | M. tuberculosis equivalent | M. avian subsp. paratuberculosis gene name | Putative functionb |
|---|---|---|---|---|---|---|---|
| 1 | 1625179 | 1633227 | 8,049 | Par MAP1484c | Rv3161c | Putative dioxygenases | |
| MAP1485c | Rv0214 | Acyl-CoA synthase | |||||
| MAP1486c | Rv0456c | Enoyl-CoA hydratase/isomerase superfamily | |||||
| MAP1487c | Rv2496c | Pyruvate dehydrogenase E1 component (beta) subunit | |||||
| MAP1488c | Rv2497c | Pyruvate dehydrogenase E1 component (alpha) subunit | |||||
| MAP1489c | Rv2750 | Putative dehydrogenase | |||||
| MAP1490 | Alpha-methylacyl-CoA racemase | ||||||
| Par MAP1491 | Alpha-methylacyl-CoA racemase | ||||||
| 2 | 1888735 | 1908664 | 19,930 | MAP1728c | yfnB | 2-Haloalkanoic acid dehalogenase | |
| MAP1729c | Rv2605c | Thioesterase II | |||||
| MAP1730c | Putative ATP/GTP-binding protein | ||||||
| MAP1731c | Hypothetical protein | ||||||
| MAP1732c | Rv0302 | Transcriptional regulator (TetR/AcrR family) | |||||
| MAP1733 | Proline-rich protein precursor | ||||||
| MAP1734 | Rv2123 | PPE family protein | |||||
| MAP1735 | Rv0217c | lipW_1 | Probable esterase | ||||
| MAP1736 | Putative tetR family transcriptional regulator | ||||||
| MAP1737 | Rv0677c | mmpS5 | Conserved small-membrane protein | ||||
| MAP1738 | Rv0676c | mmpL5 | Conserved large-membrane protein | ||||
| MAP1739c | Rv2002 | fabG3_1 | 3-Oxoacyl-(ACP) reductase | ||||
| MAP1740c | Rv3132c | Sensor histidine kinase | |||||
| MAP1741c | Rv2005c | Conserved hypothetical protein | |||||
| MAP1742c | Rv2026c | Conserved hypothetical protein | |||||
| MAP1743c | Rv2032 | Conserved hypothetical protein | |||||
| MAP1744 | Hypothetical protein | ||||||
| 3 | 2608297 | 2609525 | 1,229 | MAP2325 | Rv2416c | Enhanced intracellular survival protein |
Par, partial.
CoA, coenzyme A; ACP, acyl carrier protein.
PCR assays were then designed to bridge each of the deleted regions in the S strain (primers 75 to 80) (Table S2 in the supplemental material). The amplified product from each reaction was sequenced to determine the exact size and location for each S strain deletion with reference to the K10 genome (Table 1). The bridging PCR assays were then used to determine the conservation of these loci in 32 Australian field isolates of M. avium subsp. paratuberculosis. The 32 isolates (isolates 5 to 36 in Table S1 in the supplemental material) included 16 well-characterized S and C strains previously examined by IS900 restriction fragment length polymorphism analysis and IS1311 PCR-restriction endonuclease analysis in an extensive epidemiological examination of Johne's disease in Australia (11). The results from these PCR assays confirmed the absence of the three deleted regions in all 16 S strains and showed that these regions were present in all 16 C strains. Further examination of these regions in S and C strains from different geographical locations and representing each of the known IS900 restriction fragment length polymorphism types is required to confirm them as true markers of the S strain.
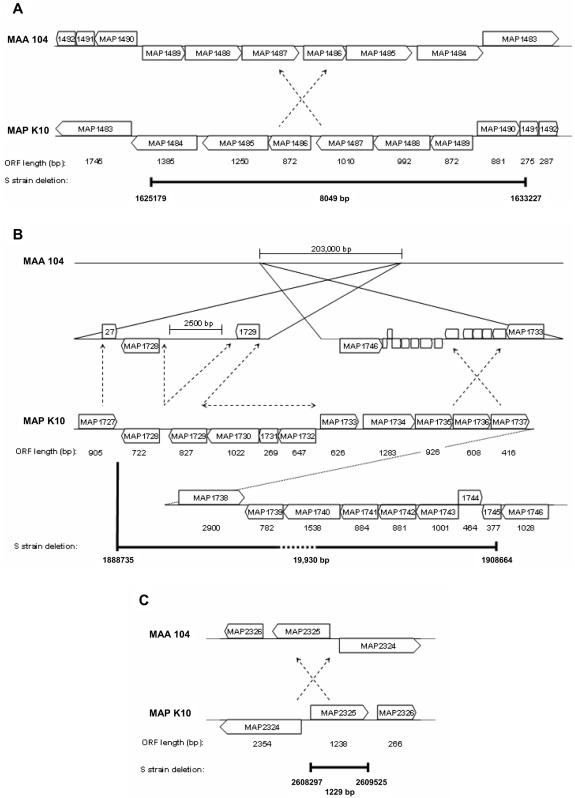
To conclude this study, the M. avium subsp. paratuberculosis K10 genome sequence (NCBI accession no. AE016958) corresponding to each of the M. avium subsp. paratuberculosis S strain deletions was used to query the M. avium subsp. avium 104 genome (The Institute for Genomic Research [TIGR]). Interestingly, deletions 1 and 3 were present but inverted in the M. avium subsp. avium 104 genome (Fig. 1A and C). However, only 17,384 bp of the 19,930 bp (∼87.2%) corresponding to deletion 2 was found in the M. avium subsp. avium 104 genome and was fragmented into two clusters separated by 203,000 bp (Fig. 1B). Clusters 1 and 2 corresponded to ORFs MAP1727 to MAP1729 and MAP1733 to MAP1746, respectively. The remaining 2,546 bp of sequence data from deletion 2 that included the ORFs corresponding to the MAP1730-MAP1732 region was not accounted for and therefore appears to be unique to the C strain of M. avium subsp. paratuberculosis. The results from this work support other studies that indicate that extensive genomic diversity exists among the members belonging to the Mycobacterium avium complex (6, 8, 9). However, they contradict the hypothesis that the S strain is an evolutionary intermediate between M. avium subsp. avium and the C strain of M. avium subsp. paratuberculosis (3) and demonstrate that further investigation is required to understand the phylogenetic and ancestral relationships of this complex.
FIG. 1.
In silico comparison of the M. avium subsp. avium (MAA) 104 genome sequence with the M. avium subsp. paratuberculosis (MAP) K10 genome sequences corresponding to the S strain deletions identified in this study. The positions of deletion 1 (A), deletion 2 (B), and deletion 3 (C) on the M. avium subsp. paratuberculosis K10 genome are displayed and mapped to the homologous sequence locations on the M. avium subsp. avium 104 genome. Deleted regions are marked by dark bars and labeled with the nucleotide positions of the M. avium subsp. paratuberculosis K10 genome flanking the deletion and with the total length of the deletion. Inversions between the two genomes are indicated by crossed arrows; however, only the regions of the inversions containing S strain deletions are displayed. The M. avium subsp. paratuberculosis K10 genome sequence with no corresponding match to the M. avium subsp. avium 104 genome is represented by a horizontal arrow.
The three deletions identified in this study represent the largest reported genomic differences found between the S and C strains of M. avium subsp. paratuberculosis to date and confirm the recent discovery of a large genomic deletion including the mmpL5 gene in the S strain of M. avium subsp. paratuberculosis (5). In total, 29,208 bp of deleted DNA, equivalent to ∼0.6% of the M. avium subsp. paratuberculosis K10 genome and including 24 complete ORFs and 2 partial ORFs, was found to be missing from the S strain genome. While putative functions have been assigned to the majority of these genes (Table 1), it remains unclear what effect the presence or absence of these genes may have on the S and C strain phenotypes. Interestingly, a number of studies have addressed this issue with orthologs from Mycobacterium tuberculosis for several of the genes identified within the S strain deletions, particularly those identified in deletion 2, and these have been discussed elsewhere (5). The results from these studies may provide valuable assistance in identifying the functions of these genes in M. avium subsp. paratuberculosis and guide future research efforts in confirming these functions and understanding the effects of their presence or absence in the S and C strain phenotypes.
A limitation of this study was the unidirectional nature of the comparisons between the S and C strains as a result of using arrays based on a C strain genome. Given that regions have been identified to be present in the S strain but absent from the C strain using subtractive hybridization techniques (3), the results from this study indicate that a thorough investigation of the C strain would benefit from a similar type of study using arrays based on the S strain. This would necessitate sequencing the entire S strain genome such that the appropriate microarrays representing all sequences present in both the S and C strains could be constructed.
Supplementary Material
Acknowledgments
This work was funded by Meat and Livestock Australia. Development of the microarray was funded by a USDA-CSREES JDIP grant subcontracted to J.P.B. and M.L.P.
We thank Kathy Granger, Sandra McKean, Leslie Reddacliff, Shayne Fell, Sue Austin, and Anna Waldron for their assistance during this study.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Collins, D. M., M. De Zoete, and S. M. Cavaignac. 2002. Mycobacterium avium subsp. paratuberculosis strains from cattle and sheep can be distinguished by a PCR test based on a novel DNA sequence difference. J. Clin. Microbiol. 40:4760-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1990. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 28:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dohmann, K., B. Strommenger, K. Stevenson, L. de Juan, J. Stratmann, V. Kapur, T. J. Bull, and G. F. Gerlach. 2003. Characterization of genetic differences between Mycobacterium avium subsp. paratuberculosis type I and type II isolates. J. Clin. Microbiol. 41:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh, I. B., and R. J. Whittington. 2005. Deletion of an mmpL gene and multiple associated genes from the genome of the S strain of Mycobacterium avium subsp. paratuberculosis identified by representational difference analysis and in silico analysis. Mol. Cell. Probes 19:371-384. [DOI] [PubMed] [Google Scholar]
- 6.Paustian, M. L., V. Kapur, and J. P. Bannantine. 2005. Comparative genomic hybridizations reveal genetic regions within the Mycobacterium avium complex that are divergent from Mycobacterium avium subsp. paratuberculosis isolates. J. Bacteriol. 187:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavlik, I., L. Bejckova, M. Pavlas, Z. Rozsypalova, and S. Koskova. 1995. Characterization by restriction endonuclease analysis and DNA hybridization using IS900 of bovine, ovine, caprine and human dependent strains of Mycobacterium paratuberculosis isolated in various localities. Vet. Microbiol. 45:311-318. [DOI] [PubMed] [Google Scholar]
- 8.Semret, M., D. C. Alexander, C. Y. Turenne, P. de Haas, P. Overduin, D. van Soolingen, D. Cousins, and M. A. Behr. 2005. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J. Clin. Microbiol. 43:3704-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semret, M., G. Zhai, S. Mostowy, C. Cleto, D. Alexander, G. Cangelosi, D. Cousins, D. M. Collins, D. van Soolingen, and M. A. Behr. 2004. Extensive genomic polymorphism within Mycobacterium avium. J. Bacteriol. 186:6332-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittington, R., I. Marsh, E. Choy, and D. Cousins. 1998. Polymorphisms in IS1311, an insertion sequence common to Mycobacterium avium and Mycobacterium avium subsp. paratuberculosis, can be used to distinguish between and within these species. Mol. Cell. Probes 12:349-358. [DOI] [PubMed] [Google Scholar]
- 11.Whittington, R. J., A. F. Hope, D. J. Marshall, C. A. Taragel, and I. Marsh. 2000. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: IS900 restriction fragment length polymorphism and IS1311 polymorphism analyses of isolates from animals and a human in Australia. J. Clin. Microbiol. 38:3240-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.