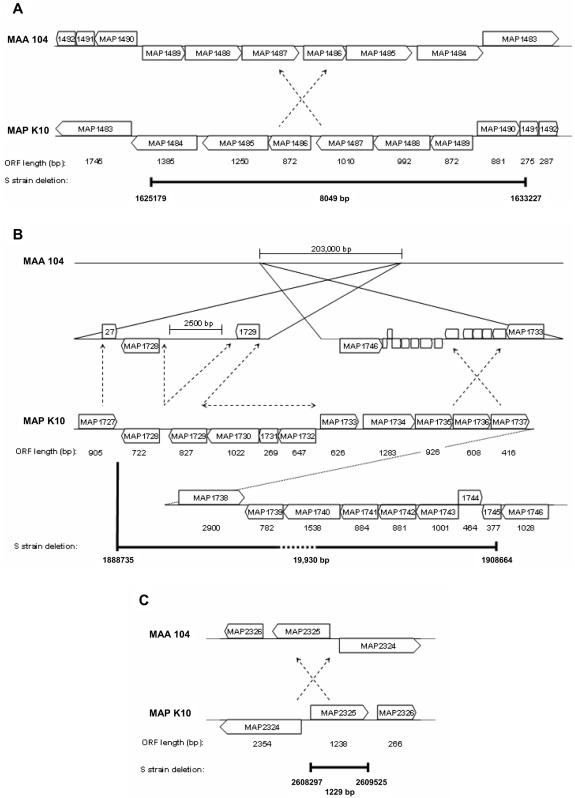
FIG. 1.
In silico comparison of the M. avium subsp. avium (MAA) 104 genome sequence with the M. avium subsp. paratuberculosis (MAP) K10 genome sequences corresponding to the S strain deletions identified in this study. The positions of deletion 1 (A), deletion 2 (B), and deletion 3 (C) on the M. avium subsp. paratuberculosis K10 genome are displayed and mapped to the homologous sequence locations on the M. avium subsp. avium 104 genome. Deleted regions are marked by dark bars and labeled with the nucleotide positions of the M. avium subsp. paratuberculosis K10 genome flanking the deletion and with the total length of the deletion. Inversions between the two genomes are indicated by crossed arrows; however, only the regions of the inversions containing S strain deletions are displayed. The M. avium subsp. paratuberculosis K10 genome sequence with no corresponding match to the M. avium subsp. avium 104 genome is represented by a horizontal arrow.