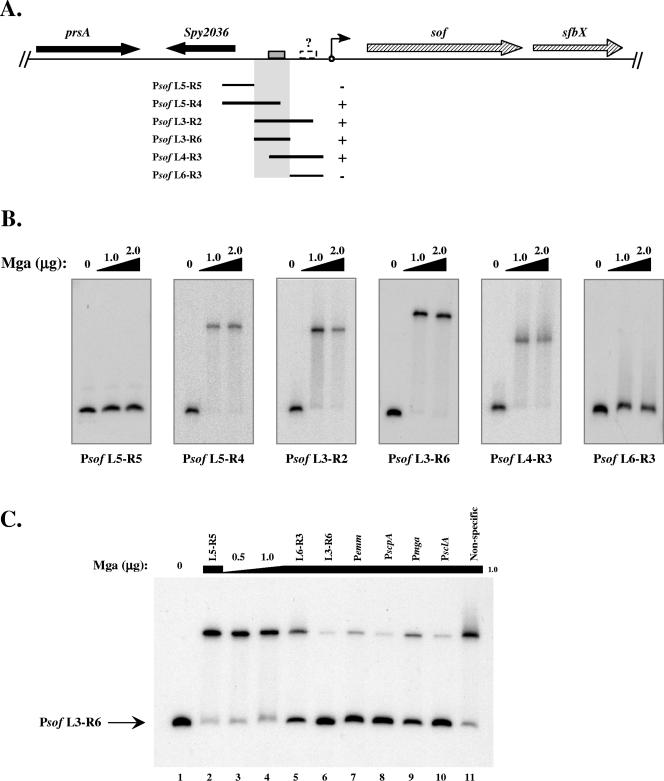
FIG. 2.
Identification of a specific Mga-binding site in the sof-sfbX promoter region. The ability of M6 Mga-His to bind to DNA within the sof-sfbX promoter was assessed by an EMSA. (A) Region surrounding sof-sfbX (hatched arrows) in the GAS genome, including a protein export protein (prsA) and a hypothetical cytosolic protein (Spy2036 in the SF370 M1 genome) (black arrows). Overlapping promoter probes in relation to the transcriptional start site of sof-sfbX (circle with arrow) are shown by the black lines. The ability of Mga-His to bind to the probes is indicated by both a plus sign and shaded region. The putative Mga-binding site (gray box) and a possible second low-affinity binding site (dashed box) are denoted on the chromosomal diagram. (B) EMSAs on Psof promoter probes (Table 2). Constant amounts (1 to 2 ng) of labeled promoter probes were incubated with increasing amounts (1.0 to 2.0 μg) of Mga-His for 15 min at 16°C prior to separation on a 5% polyacrylamide gel. (C) Radiolabeled Psof L3-R6 probe was assayed by an EMSA following incubation with 0, 0.5, and 1.0 μg Mga-His (lanes 1, 3, and 4). To the remaining binding reaction mixtures (lanes 2 and 5 to 11), 1.0 μg Mga-His and a constant amount of unlabeled competitor probe (750 ng) were added corresponding to Psof L5-R5, Psof L6-R3, Psof L3-R6, Pemm, PscpA, Pmga, PsclA, and a nonspecific rpsL fragment using the PCR primer pairs listed in Table 2.