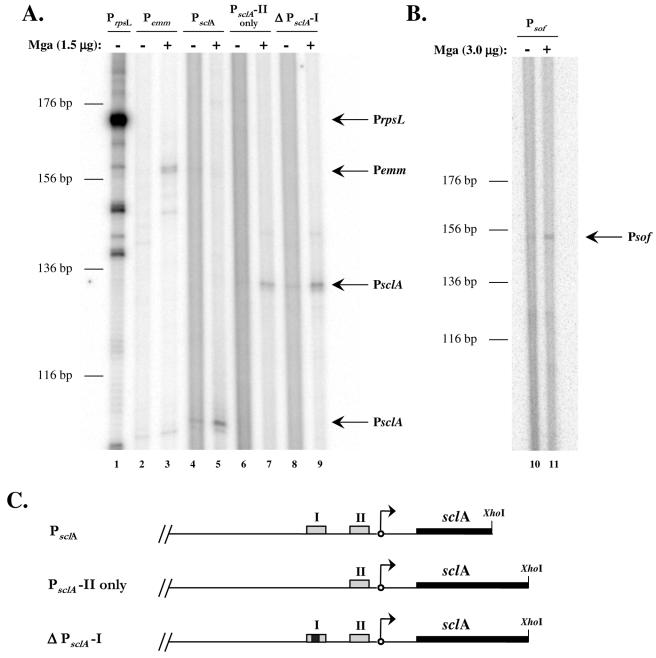
FIG. 4.
In vitro transcription of the category B Mga-regulated genes sclA and sof-sfbX. (A and B) Transcription of the sclA and sof-sfbX genes were tested in an in vitro transcription assay in the absence (−) or presence (+) of 1.5 μg of GAS-purified M6 Mga-His as described in Materials and Methods. Transcription of the Mga-independent rpsL gene and the Mga-dependent emm gene were used as controls in the assay. DNA templates (plasmid and promoter) were as follows: pKSM415, PrpsL (lane 1); pKSM416, Pemm (lanes 2 and 3); pKSM419, PsclA (lanes 4 and 5); pKSM417, PsclA-II only (lanes 6 and 7); pKSM418, ΔPsclA-I (lanes 8 and 9); and pKSM420, Psof-sfbX (lanes 10 and 11). DNA templates were incubated with purified σA, core RNA polymerase, Mga (where indicated), and a radiolabeled ribonucleotide mixture. Reaction products were ethanol precipitated and separated on a 6% denaturing polyacrylamide gel. Bands representing the transcribed products of the expected size are indicated by arrows and the name of the promoter. The sizes of the transcripts (in base pairs) were estimated on the basis of the migration of a DNA sequencing ladder. (C) Schematic of PsclA constructs including Mga-binding sites (gray boxes), deleted Mga-binding site (gray and black box), transcriptional start site (arrows), sclA (thick black lines), and location of restriction enzyme cut sites (XhoI).