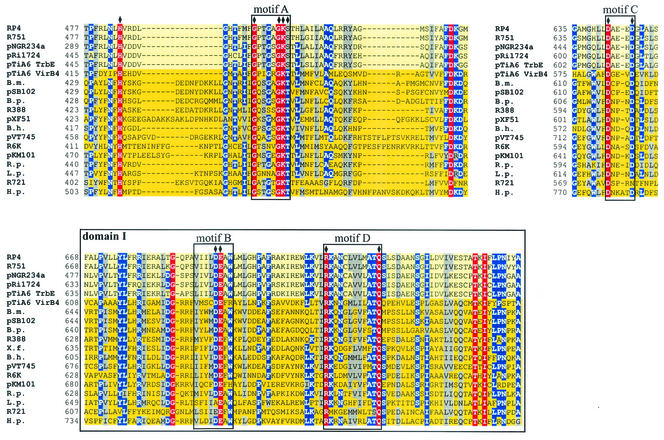
FIG. 2.
Conserved motifs in proteins belonging to the VirB4 family. Walker boxes A (motif A) and B (motif B), motifs C and D, and domain I are conserved in a multiple-sequence alignment (red, conserved residues; blue, residues conserved in more than 70% of the sequences; gray, residues with similar side chains). The subfamily of TrbE-like proteins is indicated by a light yellow background, and the subfamily of VirB4-like proteins is indicated by a dark yellow background. Amino acids in RP4 TrbE which were subjected to site-directed mutagenesis are indicated by solid diamonds above the sequences. With the exception of TrbE and VirB4 encoded by pTiA6, only the name of the plasmid or bacterium encoding the protein homologous to VirB4 is given. The following VirB4 homologs were examined (the numbers in parentheses are accession numbers): RP4, TrbE (AAA26431); R751, TrbE (NP_044243); pNGR234a, TrbEb (AAB92432); pRi1724, TrbE (BAB16246); pTiA6 TrbE (AAB95097); pTiA6 VirB4 (AAF77164); Brucella melitensis VirB4 (B.m.) (AAL53269); pSB102, TraE (CAC79179); Bordetella pertussis PtlC (B.p.) (B47301); R388, TrwK (CAC78982); pXF51, XFa0007 (AAF85576); Bartonella henselae VirB4 (B.h.) (AAF00942); pVT745, MagB03 (AAG24434); R6K, Pilx4 (CAC20141); pKM101, TraB (I79267); Rickettsia prowazekii VirB4 (R.p.) (NP_220495); Legionella pneumophila LvhB4 (L.p.) (CAB60053); R721, TraE (NP_065362); and H. pylori HP0544 (H.p.) (NP_207340).