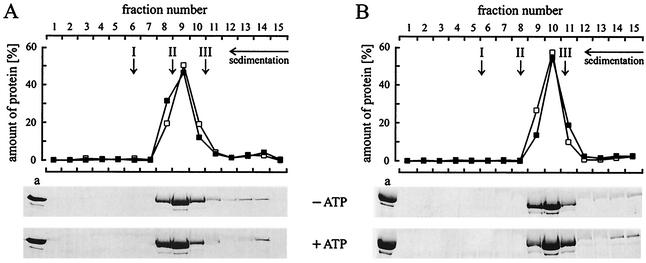
FIG. 6.
Glycerol density gradient centrifugation of RP4 TrbEHNΔ2 (A) and R388 TrwK (B). Purified proteins TrbEHNΔ2 (fraction IV, 120 μl, 0.2 mg) (Table 4) and TrwK (fraction IV, 120 μl, 0.2 mg) (Table 4) were laid on a 3.8-ml, linear, 15 to 35% (wt/vol) glycerol gradient in buffer E. Protein samples were prepared in buffer E. For analysis of conformational changes, the samples were mixed with 1 mM ATP and 10 mM MgCl2 and incubated for 10 min at 30°C prior to centrifugation. Centrifugation was carried out at 272,000 × g (rmax) for 18 h at 4°C. Fifteen fractions, each containing 15 drops of the gradient, were collected. Fifty microliters of each fraction and a 10-μl aliquot (lane a) of TrbEHNΔ2 or TrwK were separated on SDS-15% polyacrylamide gels. The gels were stained with Coomassie blue and scanned with a Personal densitometer (Molecular Dynamics). The amount of protein in each lane was quantified by using the software ImageQuant 5.0 (Molecular Dynamics). The relative amount of protein in each fraction is shown in the graphs (▪, ATP and MgCl2 added; □, ATP and MgCl2 omitted). The arrows indicate the peak positions of the reference proteins (arrow I, catalase [240 kDa, s20,w = 11.3]; arrow II, aldolase [158 kDa, s20,w = 7.8]; arrow III, BSA [66 kDa, s20,w = 4.4]).