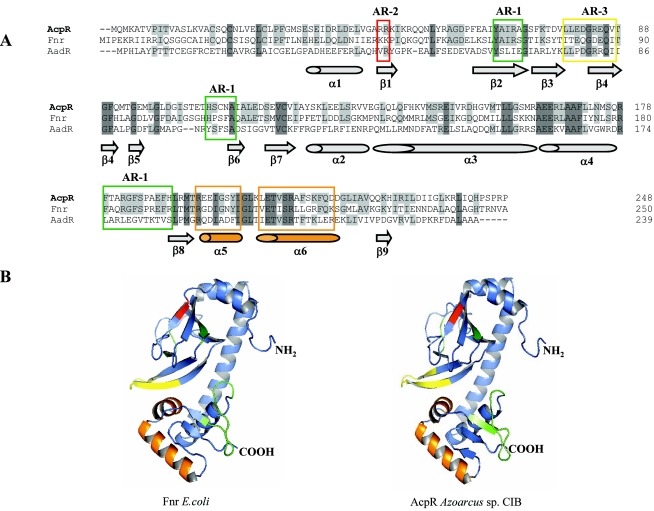
FIG.7.
Amino acid sequence alignment and three-dimensional model of AcpR. (A) Amino acid sequence alignment of AcpR, Fnr, and AadR. The accession numbers of the sequences are as follows: AcpR from Azoarcus sp. strain CIB, AY996130; Fnr from E. coli, AY629341; and AadR of R. palustris, CAE29675. The amino acid residues of each sequence are numbered on the right. Sequences were aligned using the multiple sequence alignment program Clustal (53). Amino acids are indicated by their standard one-letter codes. Dark gray represents identical residues in the three sequences. Light gray indicates identical residues in two of the three sequences. The AR-1, AR-2, and AR-3 regions are boxed in green, red, and yellow, respectively. Secondary-structure elements predicted from the AcpR three-dimensional model (B) are drawn at the bottom of the alignment. (B) Three-dimensional model of the E. coli Fnr and Azoarcus sp. strain CIB AcpR proteins; ribbon diagram showing the six α-helices (ribbons) and the nine β-strands (arrows). In green, red, and yellow are indicated the corresponding surfaces of the AR-1, AR-2, and AR-3 regions, respectively. The core helix-turn-helix fold of the winged HTH motif (2) is indicated in orange. The corresponding N and C termini are labeled NH2 and COOH, respectively.