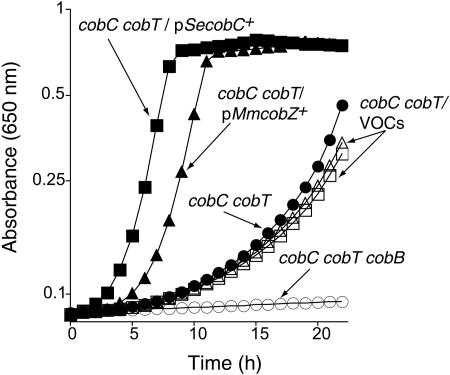
FIG. 2.
CobZ compensates for the lack of CobC during Cbl-dependent growth of a S. enterica cobC strain. Plasmid pMmcobZ+ (pCOBZ2) contained a wild-type allele of M. mazei strain Göl cobZ+ gene (ORF Mm2058; hypothetical protein, accession no. NP_634082) under the control of the T7 promoter and ribosome-binding site present in vector pT7-7 (23). The 5′ primer MM2058-5′-NdeI (5′-CATATGCATATGAAGTTATCAGATATTGAGGA-3′; restriction enzyme site is underlined) and the reverse primer MM2058-3′-BamHI (5′-CATTATGTACCGCCGGATCCTCAGTCTTTTCTTT-3′) were used to amplify a 569-bp fragment from M. mazei genomic DNA. The resulting product contained an ATG initiation codon instead of the original TTG start site. The amplified fragment was cut with NdeI/BamHI restriction enzymes, gel purified, and cloned into the NdeI/BamHI restriction site of the cloning vector pT7-7. Cbl-dependent growth was assessed in minimal medium supplemented with glucose, Cbi, and DMB. The following plasmids were introduced into strain JE2192 (cobC cobT): VOCs (vector-only controls), plasmids pT7-7 and pT7-5; plasmid pSecobC+ (pJO46); and plasmid pMmcobZ+ (pCOBZ2). Growth at 37°C with continuous shaking (19 Hz) was monitored using an EL808 UltraMicroplate Reader (BioTek Instruments). Plasmids were introduced into S. enterica by electroporation (14). S. enterica strains were grown to full density (2 × 109 CFU/ml) in nutrient broth supplemented with ampicillin (Ap; 100 μg/ml) to ensure that plasmids were maintained.