Abstract
The symbiotic pathogenic bacterium Xenorhabdus nematophila produces two distinct intracellular inclusion bodies. The pixA gene, which encodes the 185-residue methionine-rich PixA inclusion body protein, was analyzed in the present study. The pixA gene was optimally expressed under stationary-phase conditions but its expression did not require RpoS. Analysis of a pixA mutant strain showed that PixA was not required for virulence towards the insect host or for colonization of or survival within the nematode host, and was not essential for nematode reproduction. The pixA gene was not present in the genome of Xenorhabdus bovienii, which also produces proteinaceous inclusions, indicating that PixA is specifically produced in X. nematophila.
Xenorhabdus nematophila, a member of the family Enterobacteriaceae, forms a mutualistic association with the entomopathogenic nematode Steinernema carpocapsae (2, 12, 15). X. nematophila is carried in a specialized gut vesicle in the anterior portion of the intestine in the infective juvenile (IJ) form of the nematode (5). Upon invasion of the insect, the IJ enters the hemocoel and releases the bacterium into the hemolymph, where it secretes insect toxins that are involved in killing the insect host (12). In the insect cadaver, X. nematophila grows to high cell density and secretes antimicrobial and nematicidal products which protect the insect carcass from invasion by soil organisms. Xenorhabdus nematophila also produces exoenzymes that degrade insect tissues and macromolecules which contributes to the nutrient base that supports bacterial and nematode reproduction.
After several rounds of nematode reproduction, the infective juvenile form of the nematode develops, which possesses the gut vesicle that is colonized by the bacterium (5, 14). The colonized infective juvenile enters the soil environment, initiating a new life cycle upon invasion of an insect host. Each of the five established species of Xenorhabdus colonize a specific steinernematid nematode (6). These symbiotic nematode-bacterium pairs have been used as biological control agents against several agricultural pests (2).
Xenorhabdus spp. produce two distinct proteinaceous intracellular inclusions during stationary phase in culture and in insects (2, 10, 11). The sister taxon of Xenorhabdus, Photorhabdus, which engages in mutualistic associations with nematodes of the Heterorhabditidae family (1, 7, 13), also produces two morphologically distinct proteinaceous inclusion bodies (4, 8). The finding that intracellular crystalline inclusions are produced in both Xenorhabdus and Photorhabdus species suggests they are integral to the symbiotic pathogenic life cycles of these bacteria.
In X. nematophila, one inclusion body is composed of the acidic 26-kDa IP1 protein that possesses a high content of methionine residues (∼8%), while the second inclusion is composed of the neutral 22-kDa IP2 protein which is not rich in methionine residues. Together, IP1 and IP2 represent >40% of the total cellular protein in stationary-phase cells (11). Immunodetection studies showed that IP1 was present in X. nematophila but not in other Xenorhabdus species (11).
The role that crystalline inclusion body proteins play in the life cycle of Xenorhabdus spp. is not presently known. In Photorhabdus luminescens, the type 1 crystalline inclusion is composed of the 11.3-kDa CipB protein, while the type 2 crystalline inclusion is composed of the methionine-rich 11.6-kDa CipA protein (4). Strains in which either cipA or cipB was inactivated displayed a pleiotropic phenotype and were incapable of supporting nematode growth in vitro but were still virulent towards the insect host (4). Whether the absence of CipA or CipB production per se or the loss of numerous phenotypic traits in the cipA and cipB strains accounted for the inability to support nematode growth remains unclear. Analysis of the genomic sequence of Photorhabdus asymbiotica (http://www.sanger.ac.uk/Projects/P_asymbiotica/) revealed it also possesses conserved cipA and cipB genes. Finally, spontaneously forming secondary variant cell types that lack numerous phenotypic traits, including inclusion body production, are formed by both Xenorhabdus spp. and Photorhabdus spp. (4, 6, 20, 21). The X. nematophila variant cells are able to support growth of S. carpocapsae, while Heterorhabditis bacteriophora nematodes are unable to grow on the secondary variant strains of P. luminescens.
In the present study the gene encoding the IP1 protein of X. nematophila, here referred to as the protein inclusion of Xenorhabdus (PixA), was sequenced and a mutant strain in which pixA was inactivated was analyzed. The pixA strain did not display a pleiotropic phenotype, was virulent towards Manduca sexta larvae, and was able to colonize and survive within the nematode.
Nucleotide sequence accession number.
The nucleotide sequences of the pixA and cob genes described in this study were deposited in GenBank and were assigned accession number AY56156.
Sequence analysis of the pixA gene.
It was shown previously that insoluble inclusion body-containing fractions of the F1 strain of X. nematophila contained high levels of PixA which could be separated from contaminating proteins by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (20). In the present study PixA derived from the F1 strain was electrotransferred to an Immobilon-P membrane (Millipore) and subsequently subjected to N-terminal amino acid sequence analysis, which identified the first 30 residues of PixA. To obtain the full-length pixA gene sequence, the 5′ end of pixA was amplified using degenerate primers (18) designed to the N-terminal sequence of PixA, followed by an arbitrary PCR approach (9).
A PCR fragment containing the pixA gene was used to isolate a pixA-bearing clone from a plasmid library derived from the ATCC 19061 type strain of X. nematophila (see below). The nucleotide sequences of pixA of the ATCC 19061 and F1 strains were found to be identical. The pixA gene encoded a protein of 185 amino acids (Fig. 1), 15 of which were methionine residues (8.1%). Eleven of the methionine residues were located in the C-terminal half of PixA. The average methionine content in genomes related to X. nematophila is 2.58% (www.tigr.org). Blastp analysis against the nonredundant protein databases revealed that PixA did not share significant sequence similarity to any known protein.
FIG. 1.

Amino acid sequence of PixA. PixA contains 185 amino acid residues. The 15 methionine residues are shown in bold.
The calculated pI of PixA was 4.76 and the A+T content of pixA was 65.8%, quite distinct from the 55% A+T content of the rest of the genome (Forst et al., unpublished). Nucleotide sequence analysis of the flanking downstream region revealed that the cob operon (cobUSCT), which encodes enzymes for cobalamine (vitamin B12) synthesis, was convergently transcribed relative to pixA. Subsequent to the completion of the above analysis, a genome sequencing project was initiated for the ATCC 19061 strain (http://xenorhabdus.org/). Analysis of the X. nematophila genome confirmed the nucleotide sequence of pixA and the existence of a single copy of pixA in the genome. Finally, the genomic sequence of another species of Xenorhabdus, X. bovienii, was recently completed (http://xenorhabdus.org/). X. bovienii produces intracellular protein inclusions, but a pixA homologue was not identified in the X. bovienii genome.
Expression of the pixA gene.
The expression of pixA at different phases of growth was analyzed by reverse transcription (RT)-PCR (Fig. 2). pixA mRNA was not detected during the early (Fig. 2, lane 1) or mid-exponential (Fig. 2, lane 2) phase and was first apparent in cells grown to the late exponential phase (Fig. 2, lane 3). The expression of pixA reached high levels in 18-h stationary-phase cells (Fig. 2, lane 4). To determine whether the stationary-phase sigma factor RpoS was required for the expression of pixA, PixA production was examined in the rpoS strain HGB151 (19). The temporal expression and level of production of PixA in the parent and rpoS strains were indistinguishable, indicating that stationary-phase expression of pixA does not require RpoS (data not shown).
FIG. 2.
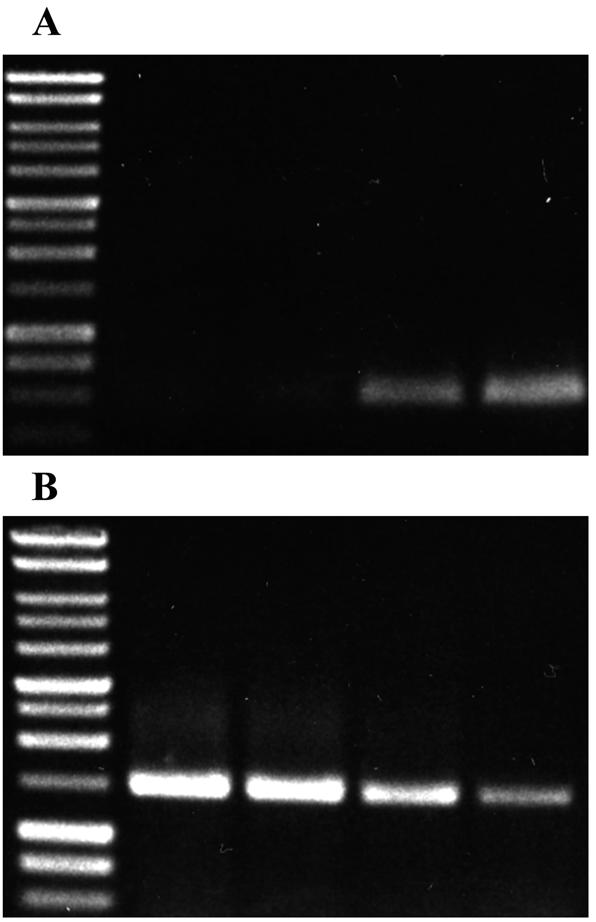
Analysis of steady-state levels of pixA mRNA by RT-PCR. (A). Total RNA was isolated from the parental X. nematophila strain at different times during growth. Lane 1, early exponential phase; lane 2, mid-exponential phase; lane 3, late exponential phase; and lane 4, stationary phase. PCR primers directed to internal sequences of pixA were used in the RT-PCR. (B). Internal control reaction using total RNA as above and primers directed to the 16S rRNA.
Insertional inactivation of pixA.
To study the role of PixA in X. nematophila, pixA was insertionally inactivated. A pST-Blue clone carrying a 400-bp PCR fragment encoding an internal region encompassing amino acids 42 to 175 of PixA was restriction digested with XbaI and PstI and the purified pixA-containing fragment was ligated into the same sites in the suicide vector pKNOCK-Cmr (3). The recombinant plasmid was electroporated into Escherichia coli S17-1 λpir and subsequently transferred by conjugation into strain ATCC 19061. Disruption of pixA in candidate mutant strains that were resistant to chloramphenicol and had the pixA-containing pKNOCK plasmid integrated into the chromosome was confirmed by Southern blot analysis. One of the confirmed strains, designated NMI1, was chosen for further analysis.
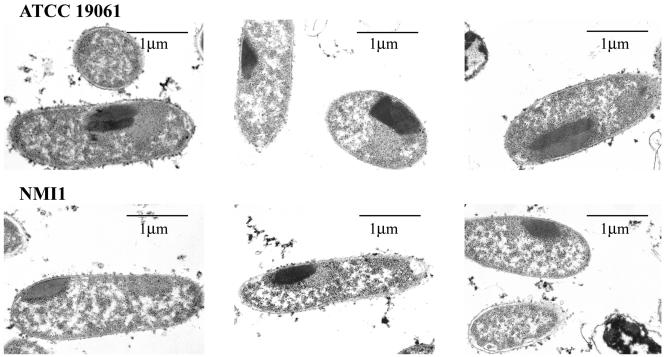
The formation of inclusion bodies in the parental and NMI1 strains was compared by transmission electron microscopy (Fig. 3). Inclusions were observed in most median longitudinal sections of the bacteria. The majority of ATCC 19061 cells examined contained two inclusion bodies (Fig. 3). The inclusions appeared to be in direct contact with each other and were not homogenous in appearance, having distinctive staining properties. These results indicated that two distinct crystalline inclusions can be produced in an individual X. nematophila cell. In contrast, only one crystalline inclusion was present in the NMI1 cells (Fig. 3).
FIG. 3.
Transmission electron microscopic analysis of inclusion bodies in the parental and NMI1 strains. Thin sections of stationary-phase cells were prepared as described in the text. Representative sections from ATCC19061 (top) and NMI1 (bottom) are shown.
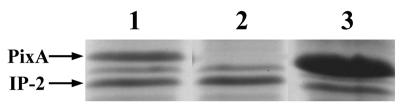
A pixA-containing low-copy-number plasmid (16, 17) was used to complement the mutant strain. SDS-polyacrylamide gel analysis showed that the parental strain produced both PixA and IP2 (Fig. 4, lane 1) while the NMI1 strain lacked PixA (Fig. 4, lane 2). PixA was found to be highly expressed in the complemented strain grown to mid-exponential phase (Fig. 4, lane 3). Early exponential growth of the complemented strain was similar to that of the parental strain. However, as the cells entered the mid-exponential phase, the growth rate was markedly reduced. This negative effect on growth of the complemented strain precluded its use in this study.
FIG. 4.
Analysis of inclusion body protein production in the parental, NMI1, and complemented NMI1 strains. Inclusion bodies isolated from either ATCC 19061 (lane 1), the NMI1 strain (lane 2), or the complemented NMI1 strain (lane 3) were analyzed by SDS-polyacrylamide gel electrophoresis.
Phenotypic characterization of the pixA strain.
The phenotypic traits examined in the NMI1 strain, such as growth rate, motility, hemolysis, lipolysis, proteolysis, and dye binding, were indistinguishable from those of the parent strain. These results were distinctly different from the cipA and cipB mutant strains of Photorhabdus luminescens, in which many of these traits were affected (4). In addition, inactivation of pixA did not affect virulence to fourth-instar Manduca sexta insects. The time at which 50% of the insect population died after injection of ∼200 cells of either the parental or NMI1 strain was 27 and 28 h, respectively.
In vitro analysis of nematode colonization.
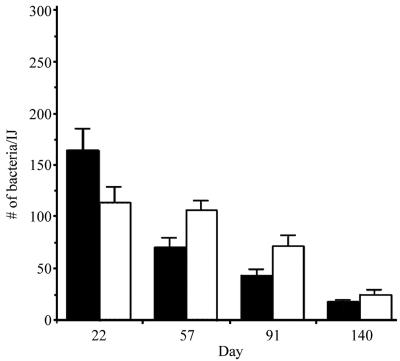
To determine whether PixA was involved in either nematode colonization or the ability of X. nematophila to survive within the intestinal vesicle, the average number of bacteria per individual live nematode colonized with either the parental or NMI1 strain was monitored over a 140-day period (Fig. 5). We added 1,000 axenic surface-sterilized IJs (20) to a bacterial lawn of either the parental or NMI1 strain. After approximately 8 to 10 days colonized IJs were collected in White traps containing tap water (22). The number of IJs produced on bacterial lawns containing either the parental or NMI1 strain was comparable, indicating that PixA was not required for nematode reproduction in vitro.
FIG. 5.
Individual colonization of IJs grown on bacterial lawns. The average number of CFU per live IJ was determined for 20 live individual nematodes at each time point indicated. The results from IJs grown on ATCC 19061 and NMI1 are shown as black and white bars, respectively.
To assess the level of colonization in individual nematodes a 1-ml aliquot of IJs was surface sterilized and resuspended in 0.5 ml of sterile LB broth. Single live nematodes were pipetted into 100 μl of LB broth in a sterile 1.5-ml microcentrifuge tube and homogenized for 70 seconds with a sterile motor-driven polypropylene pestle (Kontes). The homogenate (50 μl) was plated onto LB agar and incubated overnight. The level of colonization was determined for 20 individual IJs at each time point. At 22 days, an average of 160 and 114 CFU were recovered from nematodes grown on the parental or NMI1 strain, respectively. The CFU/IJ decreased progressively over time in both strains. With the exception of the 22-day time point, the average CFU/IJ was slightly higher for the NMI1 strain. By 140 days, an average of 9 and 12 CFU were recovered from IJs carrying the parental or NMI1 strain, respectively. These findings indicate that PixA production was not essential for either colonization of or survival in the infective juvenile.
In vivo analysis of nematode colonization.
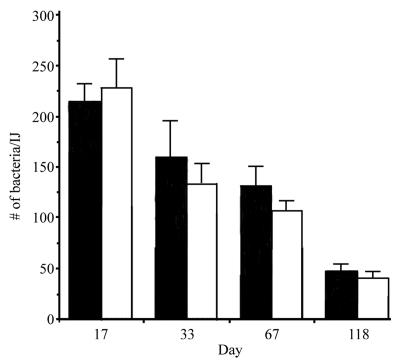
To assess whether the results obtained in the in vitro experiments accurately reflect the events that occur in vivo, the ability of the parental and NMI1 strains to colonize nematodes during natural infection of insect larvae was examined (Fig. 6). A 200-μl suspension containing 150 IJs colonized with either the parental or NMI1 strain was aliquoted into a container lined with moist filter paper. Three fourth-instar Manduca sexta larvae were then added to the container. After the insects died (24 to 36 h), they were moved to a White trap. IJs began to emerge from the insect cadaver 10 days after transfer to the White trap and were collected over a 118-day time period. IJs were surface sterilized and individual live nematodes were homogenized as described above.
FIG. 6.
Individual colonization of IJs derived from natural infections. The average number of CFU per live IJ was determined for 20 individuals at each time point indicated. The results from IJs grown on ATCC 19061 and NMI1 are shown as black and white bars, respectively.
At the 17-day time point, IJs derived from insects infected with the parental or NMI1 strain contained an average of 211 and 227 CFU/IJ, respectively. Interestingly, the level of colonization of IJs derived from infected insects was consistently greater than when the IJs were grown on bacterial lawns. By 118 days, an average of 48 and 37 CFU were recovered from IJs carrying the parental1 or NMI1 strain, respectively, indicating that PixA production was not required for either colonization of or survival within the nematode in vivo.
The above in vivo assay was used to determine whether PixA was required for nematode reproduction in the insect. M. sexta larvae were infected with IJs containing either the parental or NMI1 strain and the number of new IJs that emerged from the insect cadaver was monitored over a 20-day period. The results from five independent experiments showed that while the timing of IJ emergence varied between experiments the number of IJs produced in insects infected with the NMI1 strain was comparable to that produced in insects infected with the parental strain. Thus, nematodes are able to grow and reproduce in insects in the presence of X. nematophila organisms which lack PixA.
Finally, a competitive colonization experiment was also conducted in which a 1:1 mixture of IJs carrying either the parental or NMI1 strain was used to infect M. sexta larvae. Under these conditions, >95% of the IJs recovered were monoclonally colonized with NMI1. Almost identical results were obtained in an in vitro competitive colonization experiment in which IJs were raised on bacterial lawns containing a 1:1 mixture of the parental and NMI1 strains (data not shown). These findings suggest that X. nematophila cells that do not produce the PixA crystal protein have a selective advantage for colonization of and/or maintenance within the gut vesicle of the nematode.
Concluding remarks.
We show that the pixA gene is unique to X. nematophila and was highly expressed during stationary-phase independent of RpoS. Unlike the inactivation of cipA in Photorhabdus luminescens, inactivation of pixA did not produce a pleiotropic phenotype and the pixA strain was able to support nematode reproduction and colonization. These finding raise several intriguing questions: What is the mechanism of stationary-phase regulation of pixA? What is the nature of the differences between the inactivation of inclusion body genes in Xenorhabdus and Photorhabdus spp.? What is the function of PixA in the life cycle of X. nematophila?
Acknowledgments
We thank P. Stock, R. van der Hoeven, and H. Snyder for their critical reading of the manuscript and P. Dunn for help with statistical analysis of the colonization data.
This work was supported by the Shaw Scientist Award from the Milwaukee Foundation.
REFERENCES
- 1.Akhurst, R. J. 1980. Morphological and functional dimorphism in Xenorhabdus spp. bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. J. Gen. Microbiol. 121:303-309. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst, R. J., and N. E. Boemare. 1990. Biology and taxonomy of Xenorhabdus, p. 75-90. In R. Gaugler and H. K. Kaya (ed.), Entomopathogenic nematodes in biological control. CRC Press, Boca Raton, Fla.
- 3.Alexceyev, M. F. 1999. The pKnock series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-828. [DOI] [PubMed] [Google Scholar]
- 4.Bintrim, S. B., and J. C. Ensign. 1998. Insertional inactivation of genes encoding the crystalline inclusion proteins of Photorhabdus luminescens results in mutants with pleiotropic phenotypes. J. Bacteriol. 180:1261-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, A. F., and R. J. Akhurst. 1983., The nature of the intestinal vesicle in nematodes of the family Steinernematidae. Int. J. Parasitol. 16:511-518. [Google Scholar]
- 6.Boemare, N. E., and R. J. Akhurst. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae). J. Gen. Microbiol. 134:751-761. [Google Scholar]
- 7.Boemare, N. E., R. J. Akhurst, and R. G. Mourant. 1993. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 43:249-255. [Google Scholar]
- 8.Bowen, D. J., and J. C. Ensign. 2001. Isolation and characterization of intracellular protein inclusions produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl. Environ. Microbiol. 67:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caetano-Anolles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-94. [DOI] [PubMed] [Google Scholar]
- 10.Couche, G. A., P. R. Lehrach, R. G. Forage, G. C. Cooney, D. R. Smith, and R. P. Gregson. 1987. Occurrence of intracellular inclusions and plasmids in Xenorhabdus spp. J. Gen. Microbiol. 133:967-973. [Google Scholar]
- 11.Couche, G. A., and R. P. Gregson. 1987. Protein inclusions produced by the entomopathogenic bacterium Xenorhabdus nematophilus subsp. nematophilus. J. Bacteriol. 169:5279-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus spp. and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 13.Forst, S., and D. Clarke. 2002. Bacteria-nematode symbiosis, p. 57-78. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, United Kingdom.
- 14.Martens, E. C., K. Heugens, and H. Goodrich-Blair. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J. Bacteriol. 185:3147-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poinar, G. O. 1966. Significance of Achromobacter nematophilus Poinar and Thomas (Achromobacteraceae: Eubacteriales) in the development of the nematode, DD-136. (Neoaplectana sp. Steinernematidae). Parasitology 56:385-390. [DOI] [PubMed] [Google Scholar]
- 16.Quandt, J., and M. F. Hynes. 1983. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 17.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 18.Skanter, A, M., and L. K. Carta. 2000. Amplification of Hsp90 homologs from plant-parasitic nematodes using degenerate primers and ramped annealing PCR. BioTechniques 29:1182-1186. [DOI] [PubMed] [Google Scholar]
- 19.Vivas, E., and H. Goodrich-Blair. 2001. Xenorhabdus nematophila as a model for host-bacteria interactions: rpoS is necessary for mutualism with nematodes. J. Bacteriol. 183:4687-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volgyi, A., A. Fodor, A. Szentirmai, and S. Forst. 1998. Phase variation in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 64:1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volgyi, A., A. Fodor, and S. Forst. 2000. Inactivation of a novel gene produces a phenotypic variant cell and affects the symbiotic behavior of Xenorhabdus nematophila. Appl. Environ. Microbiol. 66:1622-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodring, J. L., and H. K. Kaya. 1988. Steinernematid and heterorhabditid nematodes: a handbook of biology and techniques. Southern Cooperative Series Bulletin 331. Arkansas Agricultural Experiment Station, Fayetteville, Ark.