Abstract
A family of genes that are likely to encode extracytoplasmic solute receptors is strongly overrepresented in several β-proteobacteria, including Bordetella pertussis. This gene family, of which members have been called bug genes, contains some examples that are contained within polycistronic operons coding for tripartite uptake transporters of the TTT family, while the vast majority are “orphan” genes. Proteomic and functional analyses demonstrated that several of these genes are expressed in B. pertussis, and one is involved in citrate uptake. The bug genes probably form an ancient family that has been subjected to a large expansion in a restricted phylogenic group.
A number of bacterial genome sequences contain large families of paralogs, genes that encode proteins of related, though not necessarily identical, functions (6, 13, 19). We have recently described a large gene family of unknown function in Bordetella pertussis, the whooping cough agent (3). These genes, named bug genes (Bordetella uptake genes; see below), code for predominantly hydrophilic proteins with predicted signal peptides. The first bug gene was identified while sequencing the regions flanking the pertussis toxin (PTX) locus and was thus called bugT (3). B. pertussis expresses its pathogenicity through the action of virulence factors coordinately regulated at the transcriptional level by the two-component regulatory system BvgAS (14, 21). Genes positively regulated by BvgAS are called vag genes (virulence-activated genes), while genes negatively regulated by BvgAS are called vrg genes (virulence-repressed genes) (8). In spite of its genomic position, bugT is not Bvg regulated, and its deletion did not affect the production or secretion of PTX (3). In contrast, another bug gene was found to correspond to the previously identified vrg24 (4).
A homology search with TBLASTN using the predicted BugT protein sequence as the query sequence (3) was performed on the genome of B. pertussis TohamaI (www.sanger.ac.uk/Projects/B_pertussis). The genes for 90 other highly similar putative proteins were found, making the bug family one of the most populated in B. pertussis (http://www.ibl.fr/articles/jbact_antoine.htm lists the sequences of the Bug proteins). The G+C percentage (67.7%) and codon usage of the Bug proteins do not significantly differ from those of other B. pertussis genes. Among the 91 copies, 12 are not functional based on the currently available genomic data, carrying large internal deletions, frameshift mutations, or the B. pertussis-specific insertion sequence IS481. The bug genes are scattered throughout the chromosome and are not systematically linked to virulence genes or insertion sequences (not shown). All 79 intact bug genes were numbered according to their position in the genome, with bugT being numbered 1 (3).
Bug genes in other bacterial species.
The genomic sequences of more than 200 bacterial genomes whose sequencing is completed or in progress were scanned using BLASTP or TBLASTN to identify bug homologs (Table 1). Bordetella bronchiseptica, Bordetella parapertussis, and a β-proteobacterium relative, Ralstonia metallidurans (formerly Alcaligenes eutrophus), are so far the only species with as many bug paralogs as B. pertussis. Several other eubacteria have between 1 and 11 bug genes, with the larger numbers found in soil-dwelling bacteria. One bug was found so far in an archaebacterium, Methanosarcina barkeri.
TABLE 1.
Predicted bug and tctBA homologs in bacterial genomes
| Bacterial species | No. of bug genes | No. of tctBA homologsa (no. of putative TTT-coding operons) |
|---|---|---|
| Agrobacterium tumefaciens | 4 | 3 (3) |
| Azotobacter vinelandii | 5 | 2 (1) |
| Bacillus halodurans | 1 | 1 (1) |
| Bordetella bronchiseptica | 181 | 4 (1) |
| Bordetella parapertussis | 143 | 4 (1) |
| Bordetella pertussis | 79 | 2 (1) |
| Burkholderia fungorum | 11 | 4 (2) |
| Corynebacterium glutamicum | 1 | 1 (1) |
| Desulfitobacterium hafniense | 5 | 5 (3) |
| Erwinia chrysanthemi | 4 | 1 (1) |
| Escherichia coli K1 | 1 | 1 (1) |
| Escherichia coli CFT073 | 1 | 1 (1) |
| Methanosarcina barkeri | 1 | 1 (1) |
| Mycobacterium smegmatis | 2 | 2 (2) |
| Pasteurella multocida | 1 | 1 (1) |
| Pseudomonas aeruginosa | 2 | 2 (2) |
| Pseudomonas fluorescens | 1 | 1 (1) |
| Pseudomonas putida KT2440 | 1 | 1 (1) |
| Pseudomonas syringae | 1 | 1 (1) |
| Ralstonia metallidurans | 102 | 5 (2) |
| Ralstonia solanacearum | 2 | 1 (1) |
| Rhodobacter sphaeroides | 3 | 3 (3) |
| Rhodopseudomonas palustris | 7 | 2 (2) |
| Salmonella enterica serover Typhimurium | 1 | 1 (1) |
| Silicibacter pomeroyi | 5 | 5 (1) |
| Sinorhizobium meliloti | 5 | 4 (4) |
| Streptomyces coelicolor | 1 | 1 (1) |
| Thermobifida fusca | 1 | 1 (1) |
| Vibrio cholerae | 1 | 1 (1) |
tctA and tctB homologs are always found within operons, usually in the order BA.
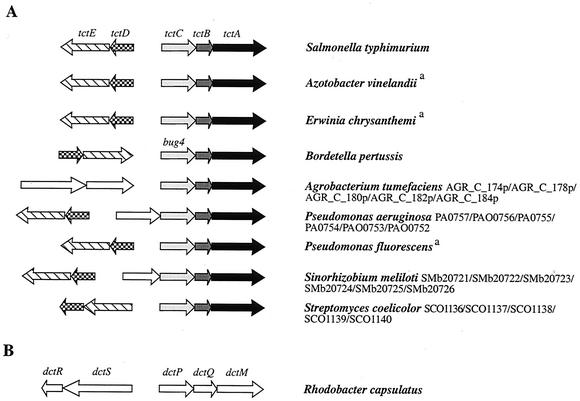
Salmonella enterica serovar Typhimurium has a gene called tctC that is homologous to bug genes (25, 27). tctC codes for a periplasmic citrate-binding protein (22) and is part of a polycistronic operon containing two other genes, tctB and tctA, that encode putative cytoplasmic membrane proteins (28) (Fig. 1). Recently, it has appeared that TctABC is the prototype of a new family of uptake transporters called tripartite tricarboxylate transporters (TTT) (www-biology.ucsd.edu/∼msaier/transport/classf.html, TC no. 2.A0.80). The TTT systems are thus the third type of transporters, in addition to the ATP-binding cassette (ABC) and the tripartite ATP-independent periplasmic (TRAP) transporters, to make use of extracytoplasmic solute receptors (ESR) (12, 23).
FIG. 1.
Organization of selected operons encoding predicted TTTs and comparison with the dctPQM operon that encodes the archetype of TRAP transporters. (A) tct operon of S. enterica serovar Typhimurium is shown as the paradigm for TTT systems. Homologs of tctC, tctB, and tctA are shown in light gray, dark gray, and black, respectively. Homologs of tctE and tctD, coding for the sensor kinase and response regulator of a two-component system involved in the regulation of tctCBA (29), are depicted as hatched and checkered arrows, respectively. Among the three predicted TTT-coding operons of Agrobacterium tumefaciens, only the one depicted here is preceded by an operon encoding a two-component system. It is not homologous to TctDE but to DctRS of Rhodobacter capsulatus, involved in the regulation of the dicarboxylate transporter operon dctPQM (see panel B). Additional genes (in white) are found in a few operons; these include genes presumably coding for a porin in Pseudomonas aeruginosa and for an ESR associated with ABC uptake transporters in Sinorhizobium meliloti. a, no accession numbers are yet available for Azotobacter vinelandii, Erwinia chrysanthemi, and Pseudomonas fluorescens. (B) The dctPQM operon that encodes the dicarboxylate TRAP transporter of Rhodobacter capsulatus (10) and the adjacent dctRS operon that encodes a two-component system are shown for comparison purposes. Although not homologous, TTT and TRAP transporters have similar molecular organizations.
All genomes containing bug/tctC homologs have between one and five operons homologous to tctBA, at least one of which includes a bug/tctC homolog (Table 1; Fig. 1). Two operons homologous to tctBA are present in B. pertussis. Only one of them forms a polycistronic operon with bug4 (Fig. 1), while the other is bicistronic and does not comprise any bug gene. In several genomes, bug/tctC homologs are more abundant than tctBA homologs, indicating the existence of “orphans” (Table 1). In B. pertussis, these orphan bug genes are either in monocistronic operons or in polycistronic operons involved in various metabolic pathways.
A functional TTT system in B. pertussis.
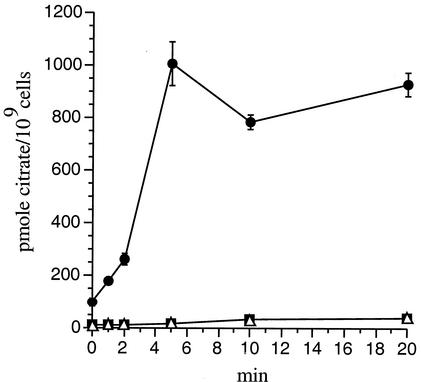
Since bug4 is the only B. pertussis bug gene in a TTT-coding operon, we investigated whether this operon is involved in citrate uptake, similar to its tctCBA relative. The following three isogenic strains were used to monitor the in vivo incorporation of radiolabeled citrate: the Tohama I derivative BPSM (16) and two mutants, one with a nonpolar in-frame deletion of bug4 and the other carrying a knockout mutation of the tctA homolog coding for the large putative membrane component of the transporter and generated by the targeted insertion of the suicide vector pFUS2 (2). The bacteria were grown in liquid modified Stainer-Scholte (MSS) medium as described previously (15). When the cultures reached optical densities of 1.5 at 600 nm, 1 mM sodium citrate (pH 7) was added, because transcriptional fusion analyses have indicated that the operon is induced by citrate (our unpublished observations). The cells were harvested 1 h later by centrifugation and resuspended in fresh MSS medium without citrate. Sixty micromolar [1,5-14C]citric acid was added (Amersham Pharmacia Biotech), and aliquots were taken after increasing labeling times. Cells were harvested by centrifugation and washed twice in MSS medium. Cell pellets were analyzed by scintillation counting. The wild-type strain incorporated citrate at a very fast rate (Fig. 2), in contrast to the mutant strains, demonstrating that both the Bug4 protein and the TctA ortholog of B. pertussis are involved in citrate uptake. Whether the low residual citrate uptake activity was mediated by another transporter has not been investigated further.
FIG. 2.
Citrate uptake by B. pertussis strains. BPSM carrying an intact copy of the operon (black circles) and two isogenic mutants, one with an in-frame deletion of the bug4 gene (black squares) and the other carrying a knockout mutation of the B. pertussis tctA ortholog (open triangles), were used to monitor the uptake of radiolabeled citrate over time. The results are expressed as picomoles of radioactive citrate incorporated by 109 cells.
Phylogenetic analyses.
Multiple alignments of 253 Bug proteins without their predicted signal peptides were performed with ClustalW and used to construct a phylogenetic tree using the PAUP software (http://www.lms.si.edu/PAUP/). This analysis revealed one small cluster of closely related Bug proteins whose genes are part of polycistronic operons involved in catechol utilization and carried on mobile genetic elements, suggesting that it has arisen from recent horizontal transfer (http://www.ibl.fr/articles/jbact_antoine.htm [Fig. A1 and Table A2]). Intriguingly, these operons did not appear to comprise genes coding for membrane components of transporters. Another cluster comprises Bug proteins whose genes are found in TTT-coding operons, including TctC of S. enterica serovar Typhimurium and a single Bug protein of B. pertussis, Bug4. The proteins of this latter cluster belong to evolutionarily divergent species, including both gram-positive and gram-negative bacteria, which suggests that they represent ancestral members. The other Bug proteins have diverged more widely. In particular, the levels of divergence between paralogs of B. pertussis or of R. metallidurans are similar to those between Bug proteins of the two species, suggesting ancient events of duplication and divergence in a common ancestor (19). Nevertheless, Bug proteins clearly form a distinct family, as evidenced by phylogenetic analyses incorporating several bona fide members of other ESR families (23; also data not shown).
The presence of a large number of orphan bug genes not included in TTT-coding operons in Bordetella and R. metallidurans remains to be explained. It is possible that several Bug proteins interact with the same set of TctAB homologs, which could represent a thrifty way to diversify transport capabilities with a minimal number of low-specificity membrane components. Alternatively, some orphan Bug proteins may have been recruited by ABC or TRAP transporters (12, 23). In addition, some Bug proteins may be involved in signal transduction, as described for other ESRs (12, 23). We have obtained preliminary evidence that Bug4 might play a role in signaling the presence of citrate (our unpublished observations).
Expression of bug genes.
To determine whether orphan bug genes are expressed in B. pertussis, a representative set was chosen for transcriptional analysis by use of lacZ fusions (2). Most genes appeared to be expressed, albeit at various levels (Table 2). No bug gene whose expression is positively regulated by BvgAS was identified. In contrast, after the addition of nicotinic acid or magnesium sulfate, which are both negative modulators of the virulence regulon, to the medium, the expression of the bug71 gene was increased approximately 2.5-fold. This indicates that the bug71 gene is a vrg, similar to bug73/vrg24 (2). The bug71 gene is found in the vicinity of the B. pertussis dermonecrotic toxin locus (3), and its ortholog in B. bronchiseptica has recently been reported to be a vrg as well (20). The biological functions of the vrg-encoded proteins in B. pertussis remain unknown, while in B. bronchiseptica several are involved in metabolic pathways or in survival under unfavorable conditions (5, 8, 20). As suggested recently, these proteins might have been relevant to nonpathogenic, free-living ancestors of present-day pathogenic Bordetella (11, 26).
TABLE 2.
β-Galactosidase activities of B. pertussis bug-lacZ recombinants
| Target gene | β-Galactosidase activity (Miller units)a in:
|
||
|---|---|---|---|
| BGb | BG + MgSO4 | BG + NA | |
| bug1 | 40 ± 1 | 33 ± 7 | 36 ± 9 |
| bug4 | 77 ± 9 | 107 ± 22 | 132 ± 27 |
| bug9 | 2,863 ± 997 | 2,639 ± 603 | 1,998 ± 240 |
| bug20 | 205 ± 39 | 292 ± 97 | 274 ± 61 |
| bug24 | <5c | <5 | <5 |
| bug43 | 411 ± 109 | 343 ± 118 | 346 ± 53 |
| bug60 | 22 ± 4 | 49 ± 5 | 31 ± 5 |
| bug64 | 85 ± 40 | 107 ± 37 | 93 ± 26 |
| bug71 | 1,193 ± 61 | 2,178 ± 292 | 2,964 ± 399 |
| bug73 | 207 ± 15 | 490 ± 50 | 496 ± 60 |
| bug79 | 323 ± 161 | 267 ± 171 | 209 ± 52 |
Data are means ± standard deviations from 3 to 5 independent experiments.
BG, Bordet Gengou medium; NA, 20 mM nicotinic acid.
Below the limit of detection.
Proteomic analyses of Bug proteins.
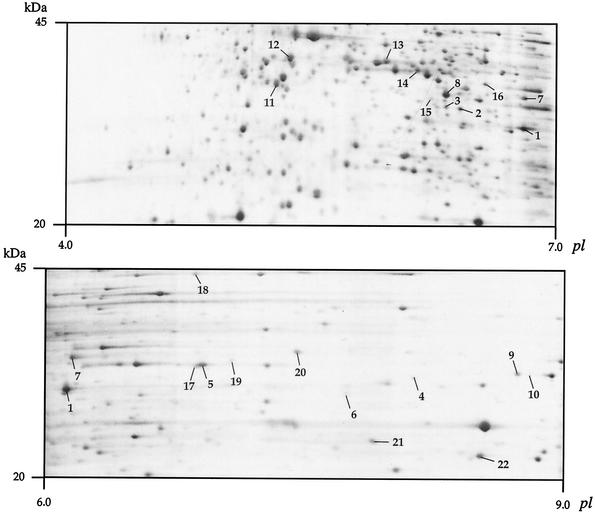
To identify Bug proteins, total protein extracts of the BPSM derivative BPDR (FHA− PTX−) (1) were analyzed by two-dimensional (2-D) electrophoresis using immobilized pH 4 to 7 or 6 to 9 gradient strips (Pharmacia). BPDR was grown in MSS medium to an optical density at 600 nm of 2. The cells were then harvested by centrifugation, and the samples for 2-D electrophoresis were prepared as described previously (24), except that the cells were broken by three passages through a French pressure cell. Proteins in the 20- to 45-kDa mass range were identified by mass fingerprinting (9) (Fig. 3). Six Bug proteins, Bug2, Bug4, Bug9, Bug20, Bug71, and Bug73, were identified. In particular, the Bug9 protein was one of the most intense protein spots in the gels. Several other abundant proteins in that region of the gel were found to correspond to predicted ESRs that belong to ABC and TRAP transporters (Fig. 3) (http://www.ibl.fr/articles/jbact_antoine.htm [Table A3] shows the sequences of these proteins).
FIG. 3.
Analyses of B. pertussis BPDR proteins by 2-D electrophoresis. The portions of pH 4 to 7 (top) and pH 6 to 9 (bottom) silver-stained gels corresponding to the 20- to 45-kDa range are shown. Only the putative periplasmic proteins involved in solute binding as identified by mass fingerprinting have been numbered (1 to 6, Bug9, -2, -73, -20, -71, and -4, respectively; 7 to 10, predicted TRAP transporter-associated ESRs; 11 to 22, predicted ABC transporter-associated ESRs). See http://www.ibl.fr/articles/jbact_antoine.htm (Table A3) for the sequences of these 22 proteins.
B. pertussis has large coding capabilities for transport systems, including approximately 100 ABC transporters and a dozen TRAP transporters (our unpublished observations), and these proteomic analyses demonstrate that it produces a number of such proteins. Generally, the global transport and metabolic capacities of an organism correlate with its ecological niche(s), and the expansion of some gene families is linked to their ability to generate physiological diversification (18, 19). The large numbers of bug genes in Bordetella and R. metallidurans indicate that they probably fulfill important functions or have done so during the evolutionary history of these bacteria. In this respect, large transport capabilities are most likely an asset to soil-dwelling R. metallidurans. In contrast, B. pertussis is a strict pathogen with more-restricted metabolic capabilities, but it is believed to have originated from free-living ancestors (11, 26). That 12 of the B. pertussis bug genes appear to be nonfunctional may indicate some genome decay, as has been observed for many genes in other pathogens (7, 17). If bug genes have evolved to bind and transport a large range of substrates for the complex metabolism of soil-dwelling microorganisms, it is plausible that some have become obsolete in the ecologically specialized, human-adapted B. pertussis.
Acknowledgments
R.A. and F.J.-D. contributed equally to this work.
We thank Julian Parkhill of the Sanger Center for his authorization to use the Bordetella genomic data prior to publication. We are grateful to P. Supply, A. Baulard, and J. Dubuisson for critical reading of the manuscript and to P. Supply for his help with phylogenetic analyses. The sequence data used in this work were obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/bp/.
H.D. is supported by the University des Sciences et Technologies de Lille I. F.J.-D. is a researcher of the CNRS. This work was supported in part by INSERM, the Institut Pasteur de Lille, and the Région Nord-Pas-de Calais.
REFERENCES
- 1.Alonso, S., K. Pethe, N. Mielcarek, D. Raze, and C. Locht. 2001. Role of ADP-ribosyltransferase activity of pertussis toxin in toxin-adhesin redundancy with filamentous hemagglutinin during Bordetella pertussis infection. Infect. Immun. 69:6038-6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoine, R., S. Alonzo, D. Raze, L. Coutte, S. Lesjean, E. Willery, C. Locht, and F. Jacob-Dubuisson. 2000. New virulence-activated and virulence-repressed genes identified by systematic gene inactivation and generation of transcriptional fusions in Bordetella pertussis. J. Bacteriol. 182:5902-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoine, R., D. Raze, and C. Locht. 2000. Genomics of Bordetella pertussis toxins. Int. J. Med. Microbiol. 290:301-305. [DOI] [PubMed] [Google Scholar]
- 4.Beattie, D. T., S. Knapp, and J. J. Mekalanos. 1990. Evidence that modulation requires sequences downstream of the promoters of two vir-repressed genes of Bordetella pertussis. J. Bacteriol. 172:6997-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie, D. T., M. J. Mahan, and J. J. Mekalanos. 1993. Repressor binding to a regulatory site in the DNA coding sequence is sufficient to confer transcriptional regulation of the vir-repressed genes (vrg genes) in Bordetella pertussis. J. Bacteriol. 175:519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, III, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, et al. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 9.Coutte, L., R. Antoine, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2001. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 18:5040-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forward, J. A., M. C. Behrendt, N. R. Wyborn, R. Cross, and D. J. Kelly. 1997. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J. Bacteriol. 179:5482-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlach, G., F. von Wintzingerode, B. Middendorf, and R. Gross. 2001. Evolutionary trends in the genus Bordetella. Microbes Infect. 3:61-72. [DOI] [PubMed] [Google Scholar]
- 12.Kelly, D. J., and G. H. Thomas. 2001. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25:405-424. [DOI] [PubMed] [Google Scholar]
- 13.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 14.Locht, C., R. Antoine, and F. Jacob-Dubuisson. 2001. Bordetella pertussis, molecular pathogenesis under multiple aspects. Curr. Opin. Microbiol. 4:82-89. [DOI] [PubMed] [Google Scholar]
- 15.Locht, C., M.-C. Geoffroy, and G. Renauld. 1992. Common accessory genes for the Bordetella pertussis filamentous hemagglutinin and fimbriae share sequence similarities with the papC and papD gene families. EMBO J. 11:3175-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menozzi, F. D., R. Mutombo, G. Renauld, C. Gantiez, J. H. Hannah, E. Leininger, M. J. Brennan, and C. Locht. 1994. Heparin-inhibitable lectin activity of the filamentous hemagglutinin adhesin of Bordetella pertussis. Infect. Immun. 62:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, et al. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 18.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier, Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 19.Saier, M. H., Jr., and I. T. Paulsen. 1999. Paralogous genes encoding transport proteins in microbial genomes. Res. Microbiol. 150:689-699. [DOI] [PubMed] [Google Scholar]
- 20.Schneider, B., D. Stubs, and R. Gross. 2002. Identification and genomic organization of gene loci negatively controlled by the virulence regulatory BvgAS two-component system in Bordetella bronchiseptica. Mol. Genet. Genomics 267:526-535. [DOI] [PubMed] [Google Scholar]
- 21.Smith, A. M., C. A. Guzman, and M. J. Walker. 2001. The virulence factors of Bordetella pertussis: a matter of control. FEMS Microbiol. Rev. 25:309-333. [DOI] [PubMed] [Google Scholar]
- 22.Sweet, G. D., C. M. Kay, and W. W. Kay. 1984. Tricarboxylate-binding proteins of Salmonella typhimurium. Purification, crystallization, and physical properties. J. Biol. Chem. 259:1586-1592. [PubMed] [Google Scholar]
- 23.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tonella, L., B. J. Walsh, J. C. Sanchez, K. Ou, M. R. Wilkins, M. Tyler, S. Frutiger, A. A. Gooley, I. Pescaru, R. D. Appel, et al. 1998. '98 Escherichia coli SWISS-2DPAGE database update. Electrophoresis 19:1960-1971. [DOI] [PubMed] [Google Scholar]
- 25.Valdivia, R. H., D. M. Cirillo, A. K. Lee, D. M. Bouley, and S. Falkow. 2000. mig-14 is a horizontally acquired, host-induced gene required for Salmonella enteritica lethal infection in the murine model of typhoid fever. Infect. Immun. 68:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Wintzingerode, F., A. Schattke, R. A. Siddiqui, U. Rosick, U. B. Gobel, and R. Gross. 2001. Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int. J. Syst. E vol. Microbiol. 51:1257-1265. [DOI] [PubMed] [Google Scholar]
- 27.Widenhorn, K. A., W. Boos, J. M. Somers, and W. W. Kay. 1988. Cloning and properties of the Salmonella typhimurium tricarboxylate transport operon in Escherichia coli. J. Bacteriol. 170:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Widenhorn, K. A., J. M. Somers, and W. W. Kay. 1988. Expression of the divergent tricarboxylate transport operon (tctI) of Salmonella typhimurium. J. Bacteriol. 170:3223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widenhorn, K. A., J. M. Somers, and W. W. Kay. 1989. Genetic regulation of the tricarboxylate transport operon (tctI) of Salmonella typhimurium. J. Bacteriol. 171:4436-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]