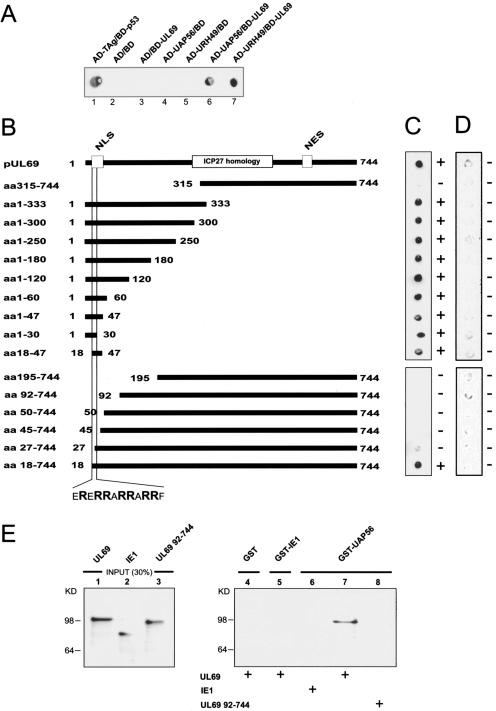
FIG. 3.
Interaction between pUL69 and cellular DEXD/H-box proteins UAP56 and URH49. (A) Yeast two-hybrid system: qualitative analysis of interactions between pUL69 and the cellular DEXD/H-box proteins UAP56 and URH49 as determined in filter lift experiments after staining for β-Gal activity. Yeast cells were transformed with two separate expression plasmids, one of which encoded either BD-UL69 or the GAL4 DNA binding domain alone (BD). The second plasmid encoded either UAP56 or URH49 as a fusion with the GAL4 activation domain (AD-UAP56 and AD-URH49) or the GAL4 activation domain alone (AD). The association of murine p53 (BD-p53) and SV40 TAg (AD-TAg) served as a positive control. (B) Mapping of the UAP56/URH49 interaction domain within pUL69. N- and C-terminal deletion mutants of pUL69 generated as in-frame fusions to the GAL4 DNA binding domain are indicated. (C) Yeast cells were transfected with a combination of vectors encoding the pUL69 deletion mutants (see panel B) and UAP56 fused to the GAL4 activation domain. Yeast colonies, selected for the presence of both plasmids, were analyzed for the expression of β-Gal by filter lift assays. (D) Qualitative analysis of the respective interaction between the various pUL69 fragments and the GAL4 activation domain alone as determined in filter lift experiments. (E) pUL69 interacts physically with UAP56 in GST pull-down experiments. In vitro-translated 35S-labeled pUL69, IE1, and an N-terminally deleted pUL69 mutant lacking the UAP56 interaction domain (UL69 aa92-744) were used for pull-down assays. Lanes 1 to 3, input protein; 30% of the amount used in the pull-down analysis is shown. Lanes 4 to 8, proteins are shown that were recovered after GST pull-down analysis. GST (lane 4), GST-IE1 (lane 5), or GST-UAP56 (lanes 6 to 8) was incubated with in vitro-translated proteins as indicated. After extensive washing, the bound proteins were resolved by SDS-PAGE, and an autoradiograph of the gel is shown.