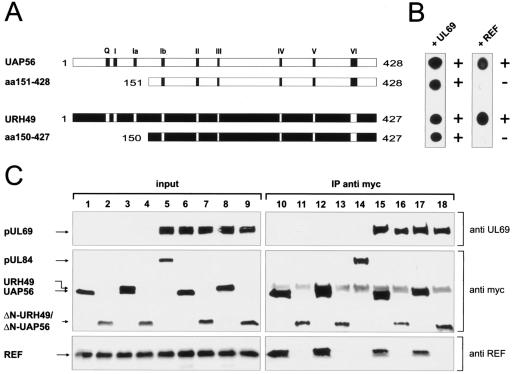
FIG. 5.
pUL69 does not rescue REF recruitment to a REF-binding-deficient UAP56 or URH49 mutant. (A) Schematic diagram illustrating N-terminal deletion mutants of UAP56 and URH49 generated as in-frame fusions to the GAL4 DNA binding domain. Conserved motifs of the DEXD/H-box family are indicated. (B) Proteins indicated in panel A were analyzed for their ability to bind GAL4 activation domain fusions of pUL69 or REF in a yeast two-hybrid assay. The experiments were performed as described in the legend to Fig. 3. (C) 293T cells were transfected with plasmids encoding myc-tagged UAP56 (lanes 1, 6, 10, and 15), myc-tagged URH49 (lanes 3, 8, 12, and 17), myc-tagged UAP56 mutant ΔN-UAP56 (lanes 2, 7, 11, and 16), myc-tagged URH49 mutant ΔN-URH49 (lanes 4, 9, 13, and 18), or a myc-UL84-expressing control plasmid (lanes 5 and 14) either in the absence (lanes 1 to 4 and 10 to 13) or in the presence (lanes 5 to 9 and 14 to 18) of a pUL69-expressing vector. The amount of protein in the input was analyzed by Western blotting (input, lanes 1 to 9). Immunoprecipitation was performed using anti-myc antibody (IP anti-myc, lanes 10 to 18). Coimmunoprecipitated proteins were visualized by Western blotting using either anti-UL69, anti-myc, or anti-REF antibodies to detect the indicated proteins. Cells cotransfected with expression plasmids encoding pUL69 and myc-tagged pUL84 (lanes 5 and 14) served as a negative control.