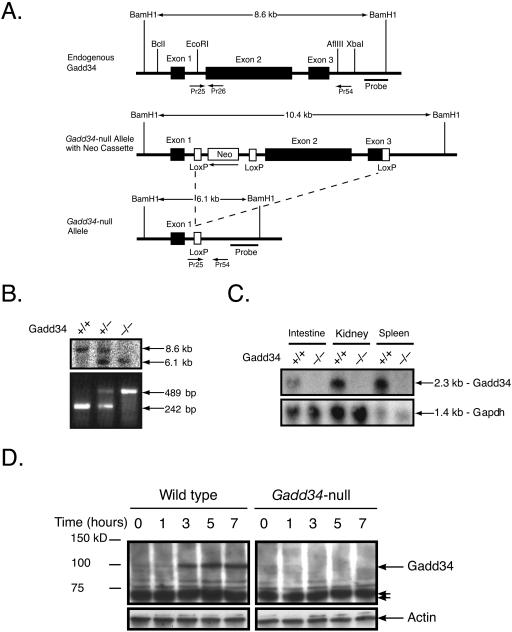
FIG. 1.
Excision of the Gadd34 coding region. (A) The Gadd34 gene was excised from the mouse genome by flanking the second and third exons (black rectangles) with LoxP (white rectangles) recombination sites. Top, a schematic representation of the Gadd34 gene in the mouse genome; middle, the Gadd34-null targeted allele containing the neomycin cassette and LoxP sites; bottom, the Gadd34 coding region deleted allele after crossing Gadd34-null targeted mice with mice expressing an eIIa-Cre recombinase transgene. (B) After crossing with the Cre recombinase transgenic mice, Gadd34-null mice were identified by genomic Southern blotting using BamHI-digested genomic DNA (top); to facilitate high-throughput genotyping, a multiple PCR approach (bottom) was used to identify the wild-type (Pr25 and Pr26) and Gadd34-null (Pr25 and Pr54) alleles. (C) Absence of Gadd34 mRNA in Gadd34-null mice was confirmed by Northern blotting. mRNA from wild-type and Gadd34-null intestine, kidney, and spleen was probed using a full-length mouse Gadd34 cDNA probe (top) and a Gapdh probe (bottom) as a loading control. (D) Wild-type and Gadd34-null early-passage embryonic day 12.5 MEFs treated with 1 μM thapsigargin and harvested at 0, 1, 3, 5, and 7 hours later. Wild-type MEFs show induction of Gadd34 by 3 hours posttreatment, with Gadd34-null MEFs showing no induction. Arrow doublets point to nonspecific bands. Actin is present as a loading control.