Abstract
Protein phosphatase 2A (PP2A) plays a prominent role in controlling accumulation of the proto-oncoprotein c-Myc. PP2A mediates its effects on c-Myc by dephosphorylating a conserved residue that normally stabilizes c-Myc, and in this way, PP2A enhances c-Myc ubiquitin-mediated degradation. Stringent regulation of c-Myc levels is essential for normal cell function, as c-Myc overexpression can lead to cell transformation. Conversely, PP2A has tumor suppressor activity. Uncovering relevant PP2A holoenzymes for a particular target has been limited by the fact that cellular PP2A represents a large heterogeneous population of trimeric holoenzymes, composed of a conserved catalytic subunit and a structural subunit along with a variable regulatory subunit which directs the holoenzyme to a specific target. We now report the identification of a specific PP2A regulatory subunit, B56α, that selectively associates with the N terminus of c-Myc. B56α directs intact PP2A holoenzymes to c-Myc, resulting in a dramatic reduction in c-Myc levels. Inhibition of PP2A-B56α holoenzymes, using small hairpin RNA to knock down B56α, results in c-Myc overexpression, elevated levels of c-Myc serine 62 phosphorylation, and increased c-Myc function. These results uncover a new protein involved in regulating c-Myc expression and reveal a critical interconnection between a potent oncoprotein, c-Myc, and a well-documented tumor suppressor, PP2A.
c-Myc is a transcription factor responsible for regulating a wide array of genes involved in cellular proliferation, growth, apoptosis, and differentiation. A number of experiments have demonstrated both the requirement for c-Myc and the importance of tightly regulating c-Myc protein levels for normal cellular function. For instance, lymphocytes and fibroblasts deleted for c-Myc cease to proliferate and exit the cell cycle (12, 64). Furthermore, homozygous deletion of the c-myc gene results in embryonic lethality in mice (11). On the other hand, sustained overexpression of c-Myc in cultured cells blocks differentiation, induces neoplastic transformation, and can initiate apoptosis when survival factors are limiting (14). A wide array of naturally occurring tumors overexpress c-Myc, due in part to chromosomal translocations, amplification, and viral insertions at the c-myc locus (8, 19). Most notably, in mice with inducible c-myc transgenes, expression of c-Myc results in neoplastic premalignant and malignant phenotypes, while withdrawal of c-Myc causes spontaneous regression of the neoplastic and malignant changes (15, 47). All of these studies highlight the importance of understanding the mechanism as well as identifying the players involved in regulating c-Myc protein levels with respect to normal and neoplastic contexts.
c-Myc expression is controlled at many levels, including gene transcription, mRNA stability, and posttranslational control of protein stability (17, 26, 29). Posttranslational regulation of c-Myc occurs through several Ras effector pathways that control a series of sequential phosphorylation events on two highly conserved residues, threonine 58 (T58) and serine 62 (S62) (56, 57, 77). These two phosphorylation sites exert opposing affects on c-Myc protein stability, with S62 phosphorylation stabilizing c-Myc and T58 phosphorylation destabilizing c-Myc. Furthermore, T58 phosphorylation requires prior S62 phosphorylation (35, 57). Upon exit from quiescence, during early G1 phase, c-Myc is stabilized by phosphorylation on S62 that can be mediated by the Ras-activated extracellular regulated kinase. Concurrent activation of phosphatidylinositol 3-kinase (PI3K) by Ras can lead to inhibition of glycogen synthase kinase-3β (GSK-3β), which is a negative regulator of c-Myc protein levels. In late G1, when PI3K activity decreases, c-Myc can become phosphorylated on T58 by active GSK-3β. This dually phosphorylated form of c-Myc associates with the phosphorylation-directed prolyl isomerase, Pin1, which can catalyze a cis-to-trans conformational change in the phospho-S62-P63 peptidyl bond of c-Myc. This form of c-Myc is then a target for protein phosphatase 2A (PP2A), which dephosphorylates S62, resulting in an unstable, singly T58-phosphorylated form of c-Myc that is a substrate for ubiquitination by SCFFbw7 and degradation by the 26S proteosome (72, 74, 77).
PP2A is a heterotrimeric protein with two common components, a structural (A) subunit and catalytic (C) subunit forming the “catalytic core,” with which a variable regulatory (B) subunit associates. To date, 25 different B subunits have been identified, which fall into four unrelated families: B, B′, B", and B‴. In total, it is estimated that there are 75 to 100 different PP2A holoenzymes, which are responsible for 30 to 50% of the total cellular serine/threonine dephosphorylation activity, depending on cell type. PP2A has been shown to be involved in regulating proliferation, growth, differentiation, and apoptosis (25). Similar to the case for c-Myc, PP2A activity is required for normal cellular function, as shown by a catalytic (Cα) subunit knockout mouse model that results in death at embryonic day 5.5 to 6 (18). However, unlike c-Myc, PP2A is generally regarded as a tumor suppressor. Global inhibition of PP2A activity results in increased cellular transformation (55). Despite the importance of PP2A as a tumor suppressor, relatively few oncogenic PP2A targets have been identified. Moreover, specific PP2A holoenzymes that target these oncoproteins have not been well described.
We now report the identification of a specific PP2A holoenzyme containing the regulatory B subunit, B56α, that associates with c-Myc. We demonstrate that the PP2A-B56α holoenzyme interacts with the trans-activation domain of c-Myc containing the S62 residue, which we previously reported to be dephosphorylated by PP2A (77). This interaction negatively regulates both c-Myc protein levels and activity. Given the potent oncogenic nature of c-Myc and the observed tumor suppressor function of PP2A, identification of the PP2A-B56α holoenzyme as a c-Myc regulatory protein adds to our understanding of two important cell regulatory pathways.
MATERIALS AND METHODS
Plasmids and RNA interference (RNAi).
Construction of expression plasmids cytomegalovirus (CMV)-empty, CMV-βgal, CMV-Myc, pCEP-small-T-antigen, pD40-His/V5-c-Myc, pD40-His/V5-c-MycT58A, and pD40-His/V5-c-MycS62A, as well as reporter constructs, E2F2-Luc, and E2F2(-E-box)-Luc, have been previously described (58, 77). pD40-His/V5-c-MycT58E and pD40-His/V5-c-MycS62D were generated using mutation primers (see the supplemental material) and TOPO cloning (Invitrogen) into pDEST40 mammalian expression vector (58, 77). pD40-His/V5-c-MycΔTAD was created by digesting CMV-Myc with PstI to remove amino acids 40 to 179, containing the transactivation domain, followed by TOPO cloning into pDEST40. CMV-HA-Myc was made by PCR amplifying the C terminus of c-Myc to include an in-frame hemagglutinin (HA) tag (see the description of primers in the supplemental material). This PCR product was used to replace the existing C terminus of c-Myc in CMV-Myc by restriction cloning using SacII and XbaI sites. pD30-PP2A-FLAG-A, pD30-PP2A-HA-C, pD40-His/V5-B55α, and pD40-His/V5-B56α were created by PCR amplification of gene coding sequences from a liver cDNA library followed by TOPO cloning into either pDEST30 mammalian expression vector (PP2A-FLAG-A and PP2A-HA-C) or pDEST40 mammalian expression vector (B55α and B56α). Expression vectors for pCEP4HA-B56α, -β, -γ1, -γ3, -δ1, and ɛ were a generous gift from David Virshup (Huntsman Cancer Institute, University of Utah). The pCAN-E4orf4 expression vector was kindly provided to us by Clodagh O'Shea (University of California, San Francisco).
Small interfering RNA (siRNA) SMARTpools for scramble control, PP2A-Cα (PPP2CA), and B55α (PPP2R2B), were purchased from Dharmacon (Lafayette, CO). Small hairpin RNA (shRNA) expression vectors were generated using OligoEngine (Seattle, WA) software to identify target sequences to each of PP2A-A, B56α, -β, -γ, -δ, and -ɛ (see the description of target sequences in the supplemental material). Oligonucleotides encoding the sense and antisense shRNA sequences were cloned into the pSUPER-shRNA expression vector (OligoEngine) by the manufacturer's protocol.
Cell lines and transfection.
HEK-293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% characterized fetal bovine serum (FBS), l-glutamine, and penicillin-streptomycin at 37°C and 5% CO2. Cells were plated to achieve 60 to 80% confluence at 24 h postsplit for transfection. SMARTpool siRNA transfections were carried out as previously described (77). All other transfections were performed using Metafectene (Biontex, Germany) according to manufacturer's specifications at a 3:1 ratio of transfection reagent to DNA (75 to 80% efficiency). Total transfected DNA (2 to 6 μg) was held constant by the addition of empty control plasmid. All transfections included 50 ng of CMV-βgal to assess transfection efficiencies between experimental conditions. Transfected cells were maintained in DMEM supplemented with 10% FBS and l-glutamine, except in shRNA experiments, in which they were maintained in 2% or 0.2% FBS and l-glutamine for the indicated time periods.
Antibodies.
The c-Myc antibodies, N262, and agarose-conjugated C-33 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). HA.11 and PP2A-A 6F9 antibodies were from Covance (Berkeley, CA). The rabbit polyclonal antibody to HA tag (rabHA) was obtained from Abcam (Cambridge, MA). The PP2A-Cα antibody was purchased from BD Biosciences (San Jose, CA). The V5 antibody was from Invitrogen (Carlsbad, CA), and the M2-FLAG and β-tubulin antibodies were from Sigma (St. Louis, MO). The c-Myc serine 62 phospho-specific antibody (αS62phospho) was generated as previously described (57). The threonine T58 phospho-specific antibody (αT58phospho) was purchased from Cell Signaling Technology (Beverly, MA), and specificity is achieved by blocking with milk (see “Western blotting and quantitation” below). The PP2A-B′α antibody was obtained from Upstate (Lake Placid, NY).
Western blotting and quantitation.
Transfected cells were lysed in 10 volumes 1.5× luciferase buffer from Promega (Madison, WI) with protease and phosphatase inhibitors (10 mM sodium fluoride, 100 mM sodium vanadate, 10 mM β-glycerolphosphate disodium pentahydrate, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 0.5 μg/ml leupeptin, 0.2 mg/ml AEBSF [4-(2-aminoethyl)benzenenesulfonyl fluoride hydrochloride], and 1.6 mg/ml iodoacetamide). Lysates were freeze-thawed three times, incubated on ice for 20 min, and subjected to β-galactosidase (β-gal) activity analysis as described previously (58). Five times sodium dodecyl sulfate sample buffer was added to a final concentration of 1.5×. Sample load volumes were adjusted by β-gal activity to account for transfection efficiency, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to Immobilon-FL (Millipore, Billerica, MA). Membranes were blocked in Odyssey Block buffer (LI-COR Biosciences, Lincoln, Nebraska), except when probed with anti-threonine 58 phospo-specific antibody, which used 5% nonfat milk in phosphate-buffered saline (PBS) for blocking. Primary antibodies were diluted in 1:1 Odyssey Block buffer-PBS, 0.05% Tween or 2.5% milk in PBS, or 0.05% Tween (P-T58 antibody) at the indicated dilutions. Primary antibodies were detected with the secondary anti-mouse and anti-rabbit near-infrared fluorescent dyes Alexa Fluor 680 (Molecular Probes, Eugene, OR) and IRDye800 (Rockland, Philadelphia, PA) used at a 1:10,000 dilution in 1:1 Odyssey Block buffer-1× PBS, 0.05% Tween or 2.5% nonfat milk in PBS, or 0.05% Tween (P-T58 antibody). Immunoblots were scanned using a LI-COR (Lincoln, Nebraska) Odyssey Infrared Imager to visualize proteins, which also allows for simultaneous anti-rabbit and anti-mouse dual-wavelength detection.
Antibody signals were quantified using LI-COR Odyssey Infrared Imager software version 1.2, which allows for linear signal quantitation over four orders of magnitude. Quantitated protein levels were normalized to control protein levels, which were set to onefold or 100% as indicated. Average protein levels and error bars representing two standard deviations were calculated from three separate experiments using Excel (Microsoft, Redmond, WA). Significant differences were also calculated from three separate experiments by t-test analysis (two-tailed distribution and two-sample unequal variance) using Excel.
Reverse transcription-PCR (RT-PCR) analysis.
Transfected HEK-293 cells were collected in 1× PBS with 1 mM EDTA, and 5% of the cells were reserved for β-gal assay and Western analysis. RNA was isolated from cells exhibiting transfection efficiencies within 5% of each other using TRIzol reagent from Invitrogen (Carlsbad, CA). cDNA was made using Moloney murine leukemia virus reverse transcriptase according to manufacturer's protocol (Invitrogen). Two times Immunomix Red from BIOLINE (Randolph, MA) was used for PCR analysis of cDNA (see the supplemental material for the primer sequence and thermocycler setup).
Cycloheximide half-life.
One-hundred-millimeter dishes of HEK-293 cells were cotransfected with 50 ng CMV-βgal, 0.5 μg pD40-His/V5-c-Myc, and 4 μg pSUPER-empty or B56α under 10% FBS conditions for 24 h. Each transfection mixture was split into six 60-mm dishes, maintained for 24 h in DMEM supplemented with 10% FBS and l-glutamine, and then starved in DMEM supplemented with 0.2% FBS and l-glutamine for 48 h. Cells were treated with 100 μg/ml cycloheximide 5 min prior to starting the indicated time course, and cells were collected at the indicated points.
Coimmunoprecipitation.
Cells were resuspended in 10 cell pellet volumes of PP2A-CoIP buffer (20 mM Tris, pH 7.5, 12.5% glycerol, 0.2% NP-40, 200 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1 mM DTT plus protease and phosphatase inhibitors). Cellular lysates were incubated on ice for 20 min and cleared by centrifugation at 20,000 × g for 10 min at 4°C. Cleared lysate volumes were adjusted for transfection efficiency by β-gal assay and incubated with either a 1:150 dilution of agarose-conjugated C-33, a 1:500 dilution of HA.11, or a 1:750 dilution of V5 antibody. Immunoprecipitates were washed three times with 10 volumes PP2A-CoIP buffer. Where specified, immunoprecipitates were more stringently washed three times with 10 volumes PP2A-CoIP buffer with 300 mM NaCl.
His-Myc-Ni-NTA columns.
Cells transfected with 6 μg pD40-His/V5-conrol, -Myc, or -MycΔTAD were resuspended in 10 cell pellet volumes Ni-nitrilotriacetic acid (Ni-NTA) lysis buffer (see the supplemental material). Cell lysates were then sonicated using a Branson Sonifier 450 sonicator (10 1-s pulses at 20% duty and output of 2), incubated on ice for 20 min, and cleared by centrifugation at 14,000 rpm for 10 min at 4°C. Cleared lysate volumes were adjusted for transfection efficiency according to β-gal activity. QIAGEN (Valencia, CA) Ni-NTA beads were added to cleared lysates and rotated for 4 hours at 4°C. Ni-NTA columns were washed three times with 10 volumes Ni-NTA CoIP buffer (see the supplemental material). Cells transfected with 6 μg pD30-PP2A-HA-C were resuspended in 10 cell pellet volumes Ni-NTA CoIP buffer, incubated on ice for 20 min, and cleared by centrifugation. Ni-NTA columns were incubated with PP2A-HA-C cleared lysate for 4 hours at 4°C and then washed three times with 10 volumes Ni-NTA wash buffer (see the supplemental material). Column-bound proteins were released by three 0.5-volume elutions with Ni-NTA elution buffer (see the supplemental material).
Luciferase assay.
Cell pellets were resuspended in 10 volumes 1.5× luciferase buffer with protease and phosphatase inhibitors, freeze-thawed three times, incubated on ice for 20 min, and cleared by centrifugation at 14,000 rpm for 10 min at 4°C. Samples were subjected to β-gal activity analysis. Luciferase activity was detected using a Promega luciferase assay kit and Berthold luminometer (Bundoora, Australia). Luciferase activity was adjusted for β-gal activity.
RESULTS
Increased PP2A activity downregulates c-Myc protein levels, dependent upon PP2A holoenzyme formation.
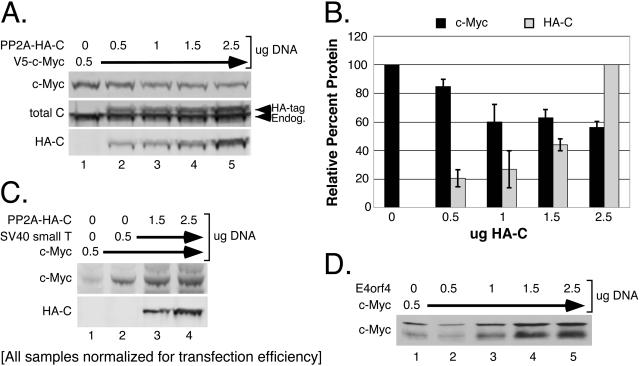
We previously demonstrated that global inhibition of PP2A strongly stabilizes c-Myc protein, allowing it to accumulate to high levels (77). These experiments relied on a variety of mechanisms for inhibiting PP2A activity, including chemical inhibition with okadaic acid, which inhibits the catalytic C subunit of PP2A (16); expression of simian virus 40 (SV40) small T antigen, which competitively inhibits regulatory B subunit binding to the structural A subunit (7, 41, 45, 62); and RNAi knockdown of the catalytic C subunit (77). To further characterize the role of PP2A in regulating c-Myc protein levels, we examined the effects of increasing cellular PP2A activity on c-Myc protein levels. We coexpressed c-Myc with increasing amounts of a stable, functional, N-terminally HA-tagged PP2A catalytic (PP2A-HA-C) subunit (2, 69). As shown in Fig. 1A and B, increasing amounts of PP2A-HA-C partially reduced c-Myc protein levels. Interestingly, the reduction was initially dose dependent (up to twofold more transfected PP2A-HA-C subunit relative to c-Myc), but no further significant decrease in c-Myc protein levels was seen with higher concentrations of PP2A-HA-C (compares lanes 1 to 3 with lanes 4 to 5 and graph).
FIG. 1.
c-Myc protein levels are negatively regulated by PP2A holoenzyme activity. (A) Increased cellular PP2A activity decreases c-Myc protein levels. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 0.5 μg pD40-His/V5-c-Myc, and increasing amounts from 0.5 μg to 2.5 μg of pD30-PP2A-HA-C, as indicated. Whole-cell lysates were collected at 36 h posttransfection, normalized for transfection efficiency by β-gal activity, and visualized by Western blot analysis with anti-V5 for c-Myc, anti-PP2A-Cα for total endogenous and ectopic PP2A-C, and anti-HA.11 for ectopic PP2A-HA-C. (B) Limited reduction of c-Myc protein levels by increased expression of PP2A-C subunit. c-Myc and PP2A-HA-C protein levels were quantified from panel A and two repeat experiments using LI-COR software (see “Western blotting and quantitation” in Materials and Methods). Average protein levels and error bars were calculated and graphed relative to the maximum level seen for c-Myc or PP2A-HA-C. (C) PP2A holoenzyme formation is required to negatively regulate c-Myc protein levels. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 0.5 μg CMV-Myc, or 0.5 μg pCEP-SV40 small T antigen, plus either 1.5 μg or 2.5 μg of pD30-PP2A-HA-C, as indicated. Whole-cell lysates were prepared and normalized as for panel A. Immunoblots were probed for c-Myc with anti-N262 and for PP2A-HA-C with anti-HA.11. (D) Adenovirus E4orf4 expression causes an increase in c-Myc protein levels. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 0.5 μg CMV-Myc and increasing amounts from 0.5 μg to 2.5 μg of pCAN-E4orf4, as indicated. Lysates were prepared and normalized as for panel A, and c-Myc protein was visualized by Western blot analysis with anti-N262.
The limited capacity of PP2A-HA-C activity to reduce c-Myc protein levels suggests that some aspect of PP2A biology is limiting. Most likely this limiting factor is related to PP2A holoenzyme formation, which involves association of the PP2A-A and -C subunits with a variable regulatory B subunit that specifies substrate recognition. This hypothesis prompted us to determine whether formation of the heterotrimeric PP2A holoenzyme was required to negatively regulate c-Myc protein levels. To test this, we inhibited PP2A holoenzyme formation by expressing SV40 small T antigen. We then coexpressed increasing amounts of PP2A-HA-C to determine whether the uncomplexed PP2A-HA-C subunit could negatively regulate c-Myc protein levels. Consistent with our results with murine fibroblasts (77), SV40 small T antigen expression increased c-Myc protein levels in human HEK-293 cells (Fig. 1C, compare lanes 1 and 2). However, addition of PP2A-HA-C no longer reduced c-Myc protein expression, at levels previously shown to maximally reduce c-Myc (Fig. 1C, lanes 3 and 4, compared to A). It is important to note that SV40 small T antigen expression reportedly does not affect PP2A-C subunit catalytic activity (76). These results strongly suggest that the PP2A-HA-C subunit must incorporate into a PP2A holoenzyme in order to negatively regulate c-Myc protein levels. Consequently, identifying the regulatory B subunit(s) that directs PP2A activity towards c-Myc is critically important to further understand the mechanism by which PP2A regulates c-Myc protein levels.
Adenovirus E4orf4 expression results in increased c-Myc protein levels.
To limit the number of potential regulatory B subunits under investigation, we made use of another viral protein, E4orf4, from adenovirus. Unlike SV40 small T antigen, E4orf4 interacts with a subset of intact PP2A holoenzymes that contain either B55α (30, 60) or B56 family members (27, 61). E4orf4 effectively inhibits these PP2A holoenzymes by redirecting their activity toward targets important for viral production (27, 60). As shown in Fig. 1D, increasing amounts of E4orf4 resulted in a dose-dependent increase in c-Myc protein levels, similar to that seen with increasing amounts of SV40 small T antigen (77). Based on this result, we narrowed our focus to B55α and B56 family members to see if any had a role in regulating c-Myc protein levels.
RNAi knockdown of the PP2A regulatory B56α subunit results in increased c-Myc protein levels.
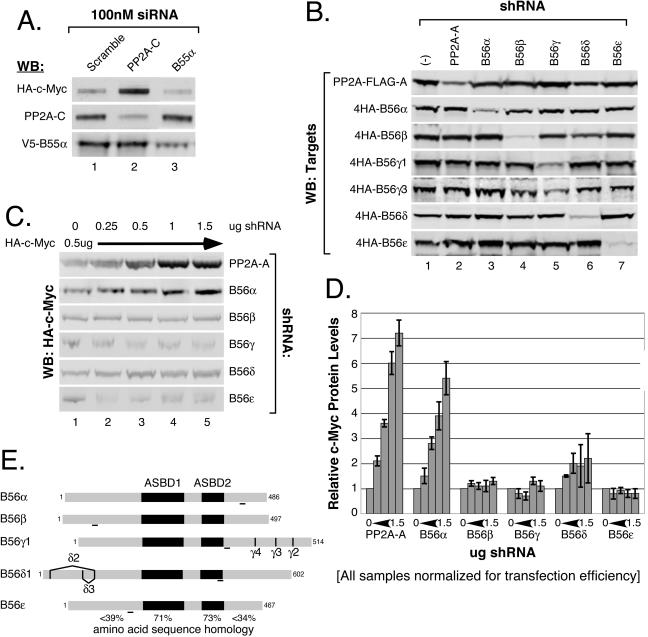
We used an RNA interference screen to individually knock down expression of B55α and B56 family regulatory subunits to assess whether they are involved in negatively regulating c-Myc protein levels. We first examined B55α by using small interfering RNA. As shown in Fig. 2A, knockdown of B55α did not cause a significant change in c-Myc protein levels compared to scramble control siRNA (compare lanes 1 and 3). In contrast, knockdown of PP2A-C resulted in a substantial increase in c-Myc protein levels (lane 2), consistent with our previous results (77). In both cases, PP2A-C and B55α were knocked down by their siRNAs (90% and 75%, respectively) (Fig. 2A, middle and bottom panels, lanes 2 and 3). We therefore concluded that B55α is not involved in negatively regulating c-Myc protein levels.
FIG. 2.
Increasing knockdown of B56α results in a robust linear increase in c-Myc protein levels. (A) siRNA knockdown of B55α does not affect c-Myc protein levels. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 0.5 μg CMV-HA-c-Myc, 0.5 μg pD40-His/V5-B55α, and siRNA (100 nM final concentration) targeted to either PP2A-C, B55α, or scramble control. Whole-cell lysates were collected at 48 h posttransfection and normalized for transfection efficiency by β-gal activity, and c-Myc, PP2A-C, and B55α proteins were visualized by Western blot (WB) analysis with anti-HA.11, anti-PP2A-Cα, and anti-V5, respectively. (B) Vector-expressed shRNAs targeted to PP2A-A and B56 family members specifically knock down intended targets. HEK-293 cells were cotransfected with 50 ng CMV-βgal; 0.5 μg of either pD30-PP2A-FLAG-A or pCEP4HA-B56α, -β, -γ1, -γ3, -δ1, or -ɛ (targets); and 2 μg pSUPER-shRNA expression vector [empty (−) or targeted to PP2A-A or B56α, -β, -γ1, -δ1, or -ɛ, as indicated]. Cells were maintained in DMEM supplemented with 2% FBS and l-glutamine for 72 h. Lysates were prepared and normalized as for panel A. Immunoblots were probed for PP2A-A with anti-M2-FLAG or for the indicated B56 family members with anti-HA.11. (C) Increasing knockdown of B56α results in increased c-Myc expression. HEK-293 cells were transfected with 50 ng CMV-βgal, 0.5 μg CMV-HA-c-Myc, and increasing amounts from 0.25 μg to 1.5 μg of pSUPER-shRNA expression vector [empty (−) or targeted to PP2A-A or B56α, -β, -γ1, -δ1, or -ɛ, as indicated]. Cells were maintained and lysates prepared and normalized as for panel B. c-Myc was visualized with anti-HA.11 by Western blot analysis. (D) Knockdown of PP2A-A or B56α results in a linear increase in c-Myc protein levels. c-Myc protein levels were quantified from panel C and two repeat experiments, and average c-Myc expression and error bars were calculated (see “Western blotting and quantitation” in Materials and Methods). Changes in c-Myc protein levels were then graphed as change relative to the shRNA control (panel C, lane 1). (E) Schematic of B56 family members. The B56 family of PP2A regulatory subunits share 71% and 73% amino acid sequence homology in the A subunit binding domains 1 and 2 (ASBD1 and -2), respectively, but significantly lower homology, less than 39% and 34%, in the N and C termini, respectively, which are believed to dictate substrate specificity (33, 39, 79). RNAi target sites used in the shRNA expression vectors are indicated by underlined regions for each B56 member. Splice variations in B56γ and B56δ are shown.
Next we examined knockdown of the B56 family of regulatory B subunits, which are encoded by five distinct genes: B56α, -β, -γ, -δ, and -ɛ (38, 40). Β56α, -β, and -ɛ each express a single isoform, whereas Β56γ and -δ express multiple isoforms due to alternative splicing, with four encoded by Β56γ and three encoded by B56δ (9, 38, 40) (see Fig. 2E for a summary). We designed vector-expressed small hairpin RNAs to knock down the PP2A structural Aα (predominant adult form) and B56α, -β, -γ, -δ, and -ɛ subunits. In the case of B56γ and -δ, the constructed shRNAs are designed to knock down all splice variants (Fig. 2E). We tested the specificities of these shRNAs by cotransfecting each shRNA construct with expression constructs for each of the targets: PP2A-A and B56α, -β, -γ1, -γ3, -δ1, or -ɛ. Each shRNA specifically reduced expression of its intended target by 70 to 90% but did not affect expression of unintended targets (Fig. 2B).
We cotransfected c-Myc with increasing amounts of each shRNA construct. As expected, increasing knockdown of the structural Aα subunit, which should result in near-global inhibition of all PP2A holoenzymes, caused a robust linear increase in c-Myc protein levels (Fig. 2C, top panel, and D). In a similar manner, increasing knockdown of B56α resulted in a linear increase in c-Myc protein levels (Fig. 2C, second panel, and 2D). In contrast, increasing knockdown of the remaining B56 subunits did not show a significant effect on c-Myc protein levels (Fig. 2C, bottom four panels, and D). Results from the E4orf4 experiment and the RNAi screen strongly suggest that B56α is likely the regulatory B subunit responsible for targeting PP2A activity towards c-Myc.
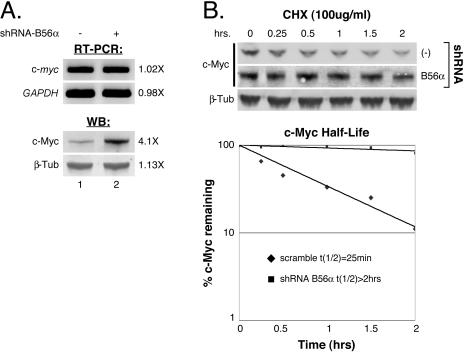
Knockdown of B56α increases c-Myc protein stability.
PP2A activity has been shown to regulate transcription (3, 42, 70), mRNA stability (28), and translation (4, 32). However, our previous data show that PP2A activity affects c-Myc protein stability (77). To determine whether PP2A-B56α might regulate c-myc transcription or mRNA stability, we examined endogenous c-myc mRNA levels by RT-PCR upon shRNA knockdown of B56α in 293 cells. c-myc mRNA levels did not change when B56α was knocked down, compared to the control (Fig. 3A, top panel). However, in the same experiment, endogenous c-Myc protein levels did increase 4.1-fold with B56α knockdown (Fig. 3A, second panel from bottom), consistent with our results from Fig. 2C and D. We next assessed whether knockdown of B56α affects c-Myc protein stability. As shown in Fig. 3B, knockdown of B56α significantly decreases the rate of c-Myc protein degradation following inhibition of protein synthesis with cycloheximide (compare middle and top panels). The quantitated increase in c-Myc half-life with B56α inhibition, from 25 min to greater than 2 h (Fig. 3B, graph), is consistent with our previous data showing a five- to sixfold increase in c-Myc stability upon global inhibition of PP2A activity (77). These results demonstrate that the increase in c-Myc protein levels upon knockdown of B56α is due to increased c-Myc protein stability rather than to increased levels or stability of c-myc mRNA.
FIG. 3.
PP2A-B56α regulates c-Myc protein stability. (A) Knockdown of B56α does not affect endogenous c-myc mRNA levels. HEK-293 cells were cotransfected with 50 ng CMV-βgal and 3 μg pSUPER-shRNA-empty (−) or pSUPER-shRNA-B56α under 10% FBS conditions for 24 h and then starved in 0.2% FBS for 48 h. Cells exhibiting transfection efficiencies by β-gal assay within 5% of each other were used for RT-PCR analysis of endogenous c-myc and GAPDH (glyceraldehyde-3-phosphate dehydrogenase gene) mRNA levels. Endogenous c-Myc and β-tubulin protein levels were also examined in the same experiment by Western blotting (WB) with anti-N262 and anti-β-tubulin, respectively. (B) c-Myc protein stability is increased upon B56α knockdown. One-hundred-millimeter dishes of HEK-293 cells were cotransfected with 50 ng CMV-βgal, 0.5 μg pD40-His/V5-c-Myc, and 4 μg pSUPER-empty or p-SUPER-B56α under 10% FBS conditions for 24 h. Each transfection mixture was split into six 60-mm dishes with 10% FBS and then starved in 0.2% FBS for 48 h. Cells were treated with 100 μg/ml cycloheximide (CHX), and cell lysates were prepared at the indicated time points after treatment. c-Myc and β-tubulin were visualized from samples at each time point with anti-V5 and anti-β-tubulin by Western analysis. c-Myc protein levels were quantified relative to β-tubulin levels and graphed as percent c-Myc protein remaining after cycloheximide treatment. Protein half-life was calculated using the Excel (Microsoft) graphing function.
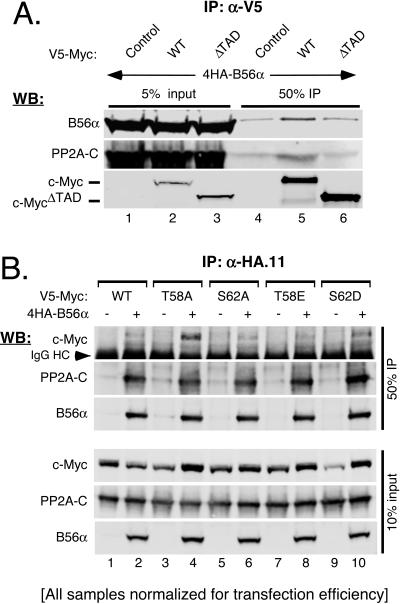
B56α associates with c-Myc.
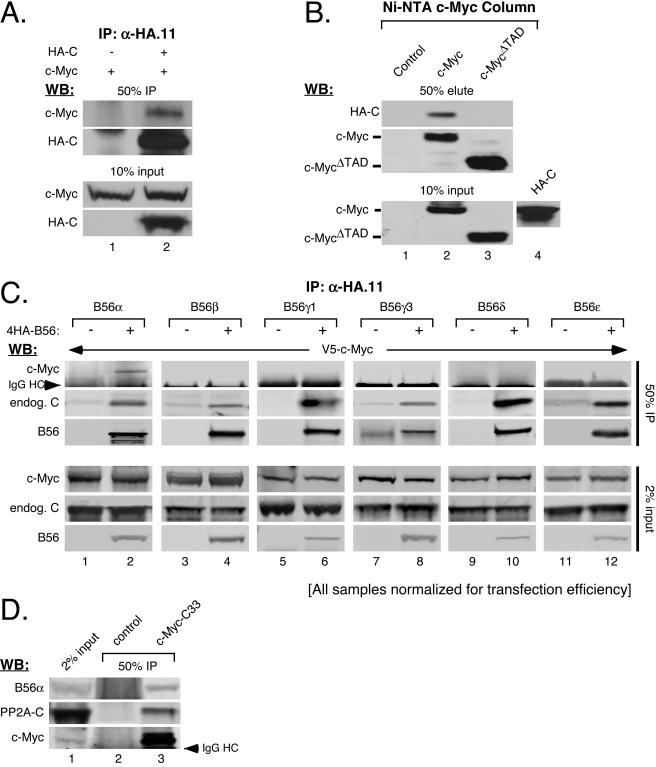
Although previous experiments presented here and elsewhere (77) have demonstrated that manipulating PP2A function can affect c-Myc protein levels, nobody has experimentally demonstrated an association between c-Myc and PP2A. We therefore initially characterized the general interaction between PP2A and c-Myc. Cells were cotransfected with an expression vector for c-Myc and either PP2A-HA-C or empty control. As shown in Fig. 4A, c-Myc coimmunoprecipitated with anti-HA antibody only in the presence of the PP2A-HA-C subunit (upper panel, compare lanes 1 and 2). We also coimmunoprecipitated PP2A-HA-C with c-Myc. His6-tagged c-Myc, c-MycΔTAD (deleted for the transactivation domain), or control Ni-NTA columns were generated and incubated with cellular lysates containing PP2A-HA-C. As shown in Fig. 4B, the PP2A-HA-C subunit was pulled out by the c-Myc column (top panel, lane 2) but not by either the control or c-MycΔTAD column (lanes 1 and 3, respectively). This not only confirms that PP2A and c-Myc associate but also shows that the interaction occurs through the N-terminal transactivation domain of c-Myc. Importantly, the transactivation domain of c-Myc contains the highly conserved S62 residue that we have previously shown to be dephosphorylated by PP2A (77).
FIG. 4.
c-Myc specifically associates with B56α. (A) c-Myc complexes with PP2A-HA-C subunit. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 3 μg CMV-Myc, and 3 μg of either pD30-PP2A-HA-C (+) or CMV-empty (−), as indicated. Cleared lysate volumes were normalized by β-gal activity and subjected to immunoprecipitation (IP) with anti-HA.11. Ten percent input volumes and 50% IP samples were analyzed by Western blotting (WB) for c-Myc with anti-N262 and for PP2A-HA-C with anti-HA.11. (B) PP2A-HA-C associates with the transactivation domain of c-Myc. HEK-293 cells cotransfected with 50 ng CMV-βgal and 3 μg either pD40-His/V5-control, -c-Myc, or -c-MycΔTAD (input shown in lanes 1 to 3 of the lower panel) were used to generate Ni-NTA-control, -c-Myc, or -c-MycΔTAD columns (see “Ni-NTA columns” in Materials and Methods). Ni-NTA columns were incubated with cleared lysates from HEK-293 cells transfected with 3 μg pD30-PP2A-HA-C (input shown in lane 4). Column-bound proteins were eluted, and 50% eluate and 10% input samples were subjected to Western blot analysis with anti-V5 for c-Myc and anti-HA.11 for PP2A-HA-C. (C) PP2A-B56α interacts with c-Myc. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 3 μg pD40-His/V5-c-Myc, and 3 μg either CMV-empty (−) or pCEP4HA-B56α, -B56β, -B56γ1, -B56γ3, -B56δ1, or -B56ɛ, as indicated. Anti-HA.11 immunoprecipitations were carried out on cleared lysates adjusted for β-gal activity. Two percent input and 50% IP samples were analyzed by Western blotting with anti-V5 for c-Myc, anti-rabHA for B56 family members, and anti-PP2A-Cα for endogenous PP2A-C. (D) Endogenous PP2A-B56α holoenzymes associate with endogenous c-Myc. Endogenous c-Myc was immunoprecipitated from HEK-293cells with agarose-conjugated C-33 antibody. Control immunoprecipitation was done using agarose A+G beads. Two percent input and 50% IP samples were subjected to Western blot analysis with anti-PP2A-B′α for endogenous B56α, anti-N262 for endogenous c-Myc, and anti-PP2A-Cα for endogenous PP2A-C subunit.
Given that c-Myc associates with PP2A, we asked whether the interaction is mediated by one or more specific regulatory B subunits. We focused on the B56 family of regulatory subunits as suggested by the E4orf4 and RNAi experiments. We coexpressed c-Myc with B56α, -β, -γ1, -γ3, -δ1, -ɛ, or -empty control and then immunoprecipitated the different B56 subunits. As shown in Fig. 4C, c-Myc clearly coimmunoprecipitated with B56α (upper panels, lane 2), but not with B56β, -γ1, -γ3, -δ1, or -ɛ (lanes 4, 6, 8, 10, and 12). Furthermore, endogenous PP2A C subunit coimmunoprecipitated with each of the B56 subunits, suggesting that PP2A holoenzymes are pulled down in each case (Fig. 4C, 2nd panel from top, even lanes). We also immunoprecipitated endogenous c-Myc from 293 cells and showed association with endogenous B56α (Fig. 4D, lane 3). Moreover, endogenous PP2A-C subunit also coimmunoprecipitated with endogenous c-Myc (middle panel, lane 3). Studies have shown that association between the regulatory B and catalytic C subunits is mediated through the structural A subunit (31, 49, 52, 63). Therefore, it is likely that intact trimeric PP2A-B56α holoenzymes associate with c-Myc through the B56α regulatory subunit.
PP2A-Β56α association with the transactivation domain of c-Myc is enhanced when serine 62 dephosphorylation is inhibited.
To determine whether B56α interacts with c-Myc near the serine 62 phosphorylation site, we immunoprecipitated wild-type c-Myc (c-MycWT) and c-MycΔTAD from 293 cells cotransfected with B56α. As shown in Fig. 5A, both B56α and endogenous PP2A-C subunit coimmunoprecipitated with c-MycWT (top two panels, lane 5) but not with c-MycΔTAD, compared to the control (lanes 4 and 6). These results demonstrate that B56α targets PP2A holoenzymes to the transactivation domain of c-Myc.
FIG. 5.
PP2A-B56α holoenzyme associates with the transactivation domain of c-Myc. (A) PP2A-B56α requires the transactivation domain of c-Myc for association. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 3 μg pCEP4HA-B56α, and 3 μg of either pD40-His/V5-contol, -c-MycWT, or -c-MycΔTAD, as indicated. Cleared lysates were collected at 36 h posttransfection and normalized for transfection efficiency by β-gal activity, and immunoprecipitations (IP) were performed with anti-V5. Five percent input and 50% IP samples were subjected to Western blot (WB) analysis with anti-V5 for c-Myc, anti-rabHA for B56α, and anti-PP2A-Cα for endogenous PP2A-C. (B) PP2A-B56α shows a stronger association with c-Myc T58A and S62D point mutants. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 3 μg of either CMV-control (−) or pCEP-4HAB56α (+), and 3 μg pD40-His/V5-c-MycWT or pD40-His/V5-c-Myc point mutants (T58A, S62A, T58E, and S62D, as indicated). Lysates were prepared and normalized as for panel A and subjected to immunoprecipitation with anti-HA.11. Immunoprecipitates were washed with a stringent buffer (see “Coimmunoprecipitation” in Materials and Methods). Ten percent input and 50% IP samples were analyzed by Western analysis with anti-V5 for c-Myc, anti-rabHA for B56α, and anti-PP2A-Cα for endogenous PP2A-C.
The transactivation domain of c-Myc contains the T58 and S62 residues, which coordinately regulate c-Myc degradation, and PP2A dephosphorylates S62. Therefore, we tested whether the phosphorylation status of T58 or S62 could affect the interaction between PP2A-B56α and c-Myc. B56α was immunoprecipitated from 293 cells coexpressing c-MycWT or c-Myc point mutants (T58A, S62A, T58E, or S62D). Immunoprecipitates were washed under stringent conditions to challenge the interaction. As shown in Fig. 5B, c-MycWT and all four point mutants coimmunoprecipitated with B56α to various degrees (top panel, even lanes). Since the S62A mutant lacks phosphorylation at both T58 and S62 (57), this result suggests that phosphorylation at T58 and/or S62 is not essential for PP2A-B56α association with c-Myc. On the other hand, both c-MycT58A and the S62 phosphorylation mimic, c-MycS62D, showed a stronger association with B56α (Fig. 5B, top panel, lanes 4 and 10, respectively). Previous studies have demonstrated that c-MycT58A shows substantially higher S62 phosphorylation levels than c-MycWT, and in vitro assays show that PP2A fails to dephosphorylate c-MycT58A (35, 57, 77). Thus, the inability of PP2A to dephosphorylate S62, as occurs with c-MycT58A and the phosphorylation mimic c-MycS62D mutants, appears to create a “substrate trap” where relatively more P-S62 c-Myc remains bound to PP2A-B56α holoenzymes.
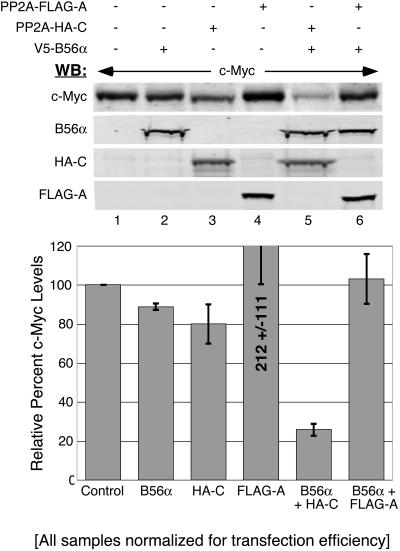
Combined expression of the catalytic and B56α subunits dramatically reduces c-Myc protein levels.
In Fig. 1, we demonstrated that increasing PP2A activity by expressing PP2A-HA-C had limited activity toward reducing c-Myc protein levels. Now that we had identified a specific PP2A holoenzyme, PP2A-B56α, that associates with c-Myc, we assessed the effects of increasing expression of this holoenzyme on c-Myc protein levels. As shown in Fig. 6, expression of either B56α or PP2A-HA-C alone showed only a slight, but consistent, reduction in c-Myc protein levels compared to the control (top panel, lanes 1 to 3, and graph). Expression of the PP2A-A subunit alone generally resulted in increased c-Myc protein levels (lane 4). Other reports have also described aberrant affects from overexpressing PP2A-A, which could explain our results (73). However, the combination of B56α with PP2A-HA-C dramatically reduced c-Myc protein levels to about 20% of that for the control (compare lanes 1 and 5 and graph). Coexpression of all three B56α, HA-tagged C, and PP2A-A subunits together also significantly reduced c-Myc protein levels to 45% (data not shown). Taken together, these results demonstrate that increasing PP2A-B56α holoenzyme levels can substantially reduce c-Myc protein expression.
FIG. 6.
Increased PP2A-B56α activity reduces c-Myc protein levels. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 1 μg CMV-Myc and 1 μg of pD30-PP2A-FLAG-A, pD30-PP2A-HA-C, and/or pD40-His/V5-B56α, as indicated. Whole-cell lysates were collected at 48 h posttransfection and normalized for transfection efficiency by β-gal activity, and samples were subjected to Western blot (WB) analysis with anti-N262 for c-Myc, anti-M2-FLAG for PP2A-A, anti-HA.11 for PP2A-C, and anti-V5 for B56α. c-Myc protein levels were quantified from three separate experiments, and average protein levels with error bars were graphed relative to control protein levels (see “Western blotting and quantitation” in Materials and Methods).
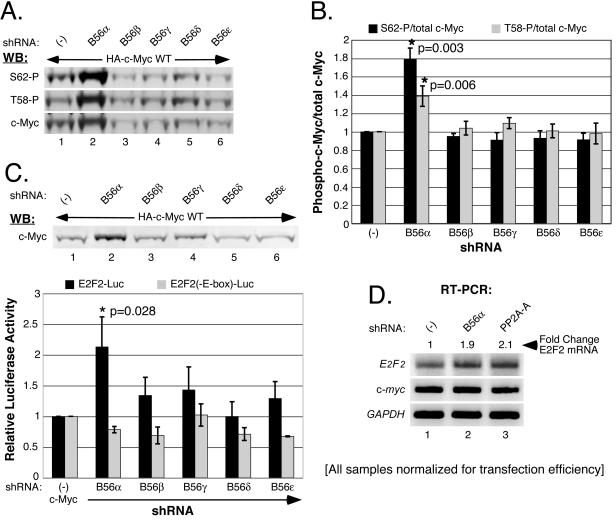
Reduced expression of B56α increases the level of serine 62-phosphorylated c-Myc.
We have previously observed that global inhibition of PP2A activity by expression of SV40 small T antigen causes an increase in S62-phosphorylated c-Myc (unpublished data). We investigated whether knockdown of B56α alone could affect the phosphorylation status of c-Myc at S62. Consistent with our previous findings, knockdown of B56α resulted in higher total c-Myc protein levels compared to the control (Fig. 7A, bottom panel, compare lanes 1 and 2). Using phospho-specific antibodies, we also observed a significant increase in both S62- and T58-phosphorylated c-Myc (top and middle panels). In contrast, knockdown of the other B56 family members did not significantly affect total c-Myc levels or S62 and T58 phosphorylation levels (Fig. 7A, lanes 3 to 6). The concurrent increase of c-Myc T58 phosphorylation with B56α knockdown is not surprising, since we are blocking the degradation of c-Myc prior to dephosphorylation on S62 by PP2A but after GSK-3β phosphorylates c-Myc on T58, and thus we are accumulating doubly phosphorylated c-Myc. By simultaneously probing for total and phospho-c-Myc, we were able to quantitate the relative increases in S62 and T58 phosphorylation with respect to total c-Myc. As shown in Fig. 7B, we observed a significant enrichment in both S62- and T58-phosphorylated c-Myc species relative to total c-Myc, with average increases of 1.8- and 1.4-fold, respectively, upon B56α knockdown. A near doubling in the population of c-Myc that is S62 phosphorylated could significantly affect c-Myc function.
FIG. 7.
Knockdown of PP2A-B56α increases S62- and T58-phosphorylated c-Myc as well as c-Myc-driven transcription. (A) c-Myc S62 and T58 phosphorylation increases upon PP2A-B56α knockdown. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 0.5 μg CMV-Myc-HA-WT, and 1.5 μg of pSUPER-shRNA expression vector [empty (−) or targeted to B56α, B56β, B56γ, B56δ, or B56ɛ, as indicated]. Whole-cell lysates were collected at 72 h posttransfection. Samples were normalized for β-gal activity and analyzed by Western blots (WB) simultaneously probed with anti-HA.11 for total c-Myc and either anti-T58phospho for T58 phosphorylation or anti-S62phospho for S62 phosphorylation. Anti-HA.11 was detected with secondary anti-mouse Alexa Fluor 680, and anti-T58phospho and anti-S62phospho were detected with secondary anti-rabbit IRDye800. The Western blots shown are representative of S62 phosphorylation, T58 phosphorylation, and total c-Myc protein levels from three separate experiments. (B) S62- and T58-phosphorylated c-Myc is enriched upon B56α knockdown. S62 or T58 phosphorylation levels along with total c-Myc levels, from simultaneously probed Western blots, were quantitated using LI-COR software (see “Western blotting and quantitation” in Materials and Methods). Ratios of S62-phosphorylation/total c-Myc and T58-phosphorylation/total c-Myc were calculated. Average ratios with error bars from the experiment shown in panel A and two repeat experiments were graphed relative to control (−) levels, and statistically significant differences are indicated (*). (C) B56α knockdown selectively increases c-Myc-driven transcription. HEK-293 cells were cotransfected with 50 ng CMV-βgal, 0.5 μg CMV-HA-c-Myc, and 10 ng of either E2F2-Luc or E2F2(-E-box)-Luc, along with 1.5 μg pSUPER-shRNA expression vector [empty (−) or targeted to B56α B56β, B56γ, B56δ, or B56ɛ, as indicated]. Whole-cell lysates were collected at 72 h posttransfection. c-Myc protein levels are shown by Western analysis with anti-HA.11. Luciferase activity was measured and corrected for transfection efficiency based on β-gal activity. Average corrected luciferase activity from three separate experiments was graphed with error bars, and statistically significant differences are indicated (*). (D) Knockdown of B56α increases endogenous E2F2 mRNA levels. HEK-293 cells were cotransfected with 50 ng CMV-βgal and 3 μg pSUPER-empty, pSUPER-B56α, or pSUPER-PP2A-A under 10% FBS conditions for 24 h and then starved in 0.2% FBS for 48 h. Cells exhibiting transfection efficiencies by β-gal assay within 5% of each other were used for RT-PCR analysis of endogenous E2F2, c-myc, and GAPDH mRNA levels.
Knockdown of PP2A-Β56α enhances c-Myc-dependent transcriptional activity.
c-Myc is a transcription factor that positively regulates the transcription of a number of target genes that are critical for cellular proliferation, growth, and differentiation (10). Consequently, we asked whether the increase in c-Myc expression observed upon knockdown of B56α has functional significance. To asses c-Myc-driven transcription, we used two luciferase reporter plasmids that contain the E2F2 promoter, which either have consensus E-box c-Myc binding sites (E2F2-Luc) or have point mutations in these binding sites that prevent c-Myc binding [E2F2(-E-box)-Luc] (58). As shown in Fig. 7C, knockdown of B56α again resulted in significantly increased c-Myc protein levels (lane 2) compared to knockdown of the other B56 family members and the shRNA control (lanes 1 and 3 to 6). Importantly, this increased accumulation of c-Myc was accompanied by a statistically significant increase in E2F2-Luc reporter activity over that seen with c-Myc when B56α is not knocked down [Fig. 7C, graph, E2F2-Luc, B56α compared to (−)]. In contrast, the negative control E2F2(-E-box)-Luc reporter plasmid, which does not support c-Myc binding, did not show a statistically significant change in activity when B56α was knocked down [Fig. 7C, E2F2(-E-box)-Luc, B56α compared to (−)]. These results demonstrate that the B56α effect on E2F2 promoter activity is c-Myc dependent. In contrast, knockdown of the remaining B56 family members showed only minor changes in the activity of either E2F2-Luc or E2F2(-E-box)-Luc (Fig. 7C, B56β, -γ, -δ, and -ɛ). We also examined the effect of knocking down B56α on endogenous c-Myc-driven transcription of the E2F2 gene. Figure 7D shows a 1.9-fold increase in E2F2 mRNA levels when B56α (lane 2) is knocked down compared to the control (lane 1). As a positive control, knockdown of PP2A-A (lane 3) resulted in a 2.1-fold increase in E2F2 mRNA levels compared to the control. The ∼2-fold increase in c-Myc-dependent E2F2 promoter activation with B56α knockdown is consistent with increases seen in c-Myc-driven transcription when c-Myc is stabilized by inhibition of PP2A activity with SV40 small T antigen (77). Based on these results, we conclude that PP2A-B56α negatively regulates c-Myc protein levels, and this level of control is important for regulating c-Myc activity. In the absence of this control, aberrant expression of functional c-Myc can occur.
DISCUSSION
Our previous work demonstrated a prominent role for PP2A in regulating c-Myc protein levels and activity (77). Given the significant number of regulatory roles assigned to PP2A, identifying the specific PP2A holoenzyme involved in regulating c-Myc gives us valuable insight into the control of c-Myc oncogenicity, as well as further understanding of how PP2A can function as a tumor suppressor. In this report, we present data identifying a specific PP2A holoenzyme containing the regulatory B subunit, B56α, that interacts with the transactivation domain of c-Myc containing the S62 residue we previously demonstrated to be dephosphorylated by PP2A. Furthermore, we show that the PP2A-B56α holoenzyme negatively regulates c-Myc protein stability and function, demonstrating the importance of this interaction with respect to c-Myc biology.
PP2A function toward c-Myc is regulated by trimeric holoenzyme formation.
It has previously been shown that ectopic expression of the PP2A-HA-C subunit in HEK-293 cells increases cellular PP2A activity (2). However, whether PP2A-HA-C requires trimeric PP2A holoenzyme formation to function has not been fully elucidated. Some studies have demonstrated that free PP2A-C and the dimeric catalytic core, PP2A-A/C, can dephosphorylate known PP2A targets in vitro (65, 76). However, other reports show that the regulatory B subunit is required for PP2A substrate specificity and greatly increases PP2A activity towards known targets (1, 6, 37, 66, 68, 75). Our findings show that in order to affect c-Myc accumulation, ectopically expressed PP2A-HA-C must incorporate into PP2A holoenzymes. Moreover, our results suggest that holoenzyme formation directed toward c-Myc is limited, presumably due to low levels or limited accessibility of the specific regulatory B subunit, B56α. Accordingly, we show that manipulation of B56α expression levels can dramatically affect c-Myc accumulation.
PP2A holoenzyme formation has been strongly implicated in cellular tumor suppressor activity. For example, there are a number of PP2A-A subunit mutations reported to occur in a variety of cancers that disrupt PP2A holoenzyme formation (50, 51, 71). Additionally, inhibition of PP2A holoenzyme assembly by expression of SV40 small T antigen has been shown to be a critical step in the experimental conversion of primary human cells to a transformed tumorigenic state (20, 45, 53, 78). Interestingly, the requirement for PP2A inhibition in this assay can be circumvented by expression of the stable c-MycT58A mutant, which is resistant to PP2A-mediated dephosphorylation and degradation (77). These findings highlight the importance of PP2A holoenzyme formation for tumor suppressor activity and reveal the critical nature of PP2A regulation of c-Myc for normal cellular function.
B56α targets c-Myc and is a key regulator of PP2A tumor suppressor activity.
The B56 family of PP2A regulatory B subunits is diverse, with each member displaying distinct tissue expression, developmental expression, and subcellular localization patterns (36, 40). In general, the B56 family members have been regarded as tumor suppressors (67). To date, a great deal of attention has focused on the B56γ isoforms. An N-terminal deletion mutant of B56γ1, found in a mouse melanoma cell line, has been shown to result in increased metastasis by deregulation of paxillin (23) and inhibition of the gamma irradiation cell cycle check point, contributing to genomic instability (23, 24). B56γ3 is reported to regulate Chk2 and p300 protein levels (6, 13), as well as to be involved in cell cycle control by regulating Mdm2 and p53, through its association with cyclin G (43, 44).
Although the B56γ subunits are clearly tumor suppressors, knockdown of the B56γ subunits recapitulates the requirement for SV40 small T antigen in cellular transformation of primary human cells only after a significantly longer period of time (7). This result suggests that other PP2A holoenzymes targeted by SV40 small T antigen play an essential role in the transformation of human cells. In this regard, our results showing that c-Myc is a target of PP2A-B56α identify another such PP2A holoenzyme targeted by SV40 small T antigen with tumor suppressor function.
The B56α regulatory subunit is reported to be the most ubiquitously expressed isoform of the B56 family (36, 40), making it an ideal regulatory B subunit to target PP2A activity towards c-Myc, since c-Myc is also ubiquitously expressed. Although there are no known mutations in B56α in human cancers, two mutations identified in the PP2A-A subunit from human tumors have been shown to specifically disrupt the binding of B56α to PP2A-A, selectively inhibiting PP2A-B56α holoenzyme formation (51). In addition, several other important cellular regulatory proteins have been shown to be targeted by B56α. The antiapoptotic protein Bcl2 has been shown to be inactivated by B56α (54), and B56α plays a crucial role in negatively regulating protein levels of the Wnt-signaling transcription factor β-catenin (34, 59). The discovery that c-Myc is a B56α target places B56α at a critical junction point for regulating multiple potent oncoproteins.
Interplay between the oncogenic activity of c-Myc and the tumor suppressor function of PP2A-B56α.
Despite its critical role in regulating multiple oncoproteins, B56α transcript levels are found to be relatively low in most tissues, and data presented here suggest that B56α protein is limiting toward c-Myc (36). Presumably, under normal or quiescent cellular conditions, when c-Myc and β-catenin proteins levels are low and Bcl2 is not activated, these low levels of B56α are sufficient. However, aberrant deregulation of any one of these oncoproteins could sequester the majority of B56α and thereby cause deregulation of the other oncoproteins regulated by PP2A-B56α. In fact, ΔNp63α, an oncogenic form of the p53 family member p63, which is overexpressed in squamous cell carcinomas, has been reported to associate with B56α, and this results in the accumulation of β-catenin (46). Our finding that c-MycT58A has a stronger association with PP2A-B56α than wild-type c-Myc could mean that it acts in a manner similar to that of ΔNp63, explaining in part the increased oncogenicity of c-MycT58A versus c-MycWT (22, 48, 57). Furthermore, c-MycT58A shows reduced apoptosis compared to c-MycWT, which could be facilitated by the efficient sequestering of PP2A-B56α by c-MycT58A, thus preventing B56α-mediated inactivation of the antiapoptotic Bcl2 (5, 21).
Supplementary Material
Acknowledgments
We thank David Virshup for the HA-tagged B56 family expression vectors and Clodagh O'Shea for the E4orf4 expression vector.
Funding for this work was provided by NIH Training Grant 5-T32-GM08617 and OHSU TL Tartar Trust Research Fellowship AMEDG0063 to Hugh K. Arnold and by NIH grants RO1-CA100855 and KO1-CA086957 to Rosalie C. Sears.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agostinis, P., R. Derua, S. Sarno, J. Goris, and W. Merlevede. 1992. Specificity of the polycation-stimulated (type-2A) and ATP, Mg-dependent (type-1) protein phosphatases toward substrates phosphorylated by P34cdc2 kinase. Eur. J. Biochem. 205:241-248. [DOI] [PubMed] [Google Scholar]
- 2.Al-Murrani, S. W., J. R. Woodgett, and Z. Damuni. 1999. Expression of I2PP2A, an inhibitor of protein phosphatase 2A, induces c-Jun and AP-1 activity. Biochem. J. 341:293-298. [PMC free article] [PubMed] [Google Scholar]
- 3.Altiok, S., M. Xu, and B. M. Spiegelman. 1997. PPARgamma induces cell cycle withdrawal: inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev. 11:1987-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andjelkovic, N., S. Zolnierowicz, C. Van Hoof, J. Goris, and B. A. Hemmings. 1996. The catalytic subunit of protein phosphatase 2A associates with the translation termination factor eRF1. EMBO J. 15:7156-7167. [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, D. W., G. F. Claassen, S. R. Hann, and M. D. Cole. 2000. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol. Cell. Biol. 20:4309-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J., J. R. St.-Germain, and Q. Li. 2005. B56 regulatory subunit of protein phosphatase 2A mediates valproic acid-induced p300 degradation. Mol. Cell. Biol. 25:525-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, W., R. Possemato, K. T. Campbell, C. A. Plattner, D. C. Pallas, and W. C. Hahn. 2004. Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell 5:127-136. [DOI] [PubMed] [Google Scholar]
- 8.Cole, M. D. 1986. The myc oncogene: its role in transformation and differentiation. Annu. Rev. Genet. 20:361-384. [DOI] [PubMed] [Google Scholar]
- 9.Csortos, C., S. Zolnierowicz, E. Bako, S. D. Durbin, and A. A. DePaoli-Roach. 1996. High complexity in the expression of the B′ subunit of protein phosphatase 2A0. Evidence for the existence of at least seven novel isoforms. J. Biol. Chem. 271:2578-2588. [DOI] [PubMed] [Google Scholar]
- 10.Dang, C. V., L. M. Resar, E. Emison, S. Kim, Q. Li, J. E. Prescott, D. Wonsey, and K. Zeller. 1999. Function of the c-Myc oncogenic transcription factor. Exp. Cell Res. 253:63-77. [DOI] [PubMed] [Google Scholar]
- 11.Davis, A. C., M. Wims, G. D. Spotts, S. R. Hann, and A. Bradley. 1993. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7:671-682. [DOI] [PubMed] [Google Scholar]
- 12.de Alboran, I. M., R. C. O'Hagan, F. Gartner, B. Malynn, L. Davidson, R. Rickert, K. Rajewsky, R. A. DePinho, and F. W. Alt. 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14:45-55. [DOI] [PubMed] [Google Scholar]
- 13.Dozier, C., M. Bonyadi, L. Baricault, L. Tonasso, and J. M. Darbon. 2004. Regulation of Chk2 phosphorylation by interaction with protein phosphatase 2A via its B′ regulatory subunit. Biol. Cell. 96:509-517. [DOI] [PubMed] [Google Scholar]
- 14.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 15.Felsher, D. W., and J. M. Bishop. 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 4:199-207. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez, J. J., M. L. Candenas, M. L. Souto, M. M. Trujillo, and M. Norte. 2002. Okadaic acid, useful tool for studying cellular processes. Curr. Med. Chem. 9:229-262. [DOI] [PubMed] [Google Scholar]
- 17.Flinn, E. M., C. M. Busch, and A. P. Wright. 1998. Myc boxes, which are conserved in Myc family proteins, are signals for protein degradation via the proteasome. Mol. Cell. Biol. 18:5961-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotz, J., A. Probst, E. Ehler, B. Hemmings, and W. Kues. 1998. Delayed embryonic lethality in mice lacking protein phosphatase 2A catalytic subunit Calpha. Proc. Natl. Acad. Sci. USA 95:12370-12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory, M. A., and S. R. Hann. 2000. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol. Cell. Biol. 20:2423-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn, W. C., S. K. Dessain, M. W. Brooks, J. E. King, B. Elenbaas, D. M. Sabatini, J. A. DeCaprio, and R. A. Weinberg. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22:2111-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemann, M. T., A. Bric, J. Teruya-Feldstein, A. Herbst, J. A. Nilsson, C. Cordon-Cardo, J. L. Cleveland, W. P. Tansey, and S. W. Lowe. 2005. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henriksson, M., A. Bakardjiev, G. Klein, and B. Luscher. 1993. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene 8:3199-3209. [PubMed] [Google Scholar]
- 23.Ito, A., T. R. Kataoka, M. Watanabe, K. Nishiyama, Y. Mazaki, H. Sabe, Y. Kitamura, and H. Nojima. 2000. A truncated isoform of the PP2A B56 subunit promotes cell motility through paxillin phosphorylation. EMBO J. 19:562-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, A., Y. Koma, K. Watabe, T. Nagano, Y. Endo, H. Nojima, and Y. Kitamura. 2003. A truncated isoform of the protein phosphatase 2A B56gamma regulatory subunit may promote genetic instability and cause tumor progression. Am. J. Pathol. 162:81-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssens, V., and J. Goris. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353:417-439.11171037 [Google Scholar]
- 26.Jones, T. R., and M. D. Cole. 1987. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol. Cell. Biol. 7:4513-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanopka, A., O. Muhlemann, S. Petersen-Mahrt, C. Estmer, C. Ohrmalm, and G. Akusjarvi. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185-187. [DOI] [PubMed] [Google Scholar]
- 28.Keen, J. C., Q. Zhou, B. H. Park, C. Pettit, K. M. Mack, B. Blair, K. Brenner, and N. E. Davidson. 2005. Protein phosphatase 2A regulates estrogen receptor alpha (ER) expression through modulation of ER mRNA stability. J. Biol. Chem. 280:29519-29524. [DOI] [PubMed] [Google Scholar]
- 29.Kelly, K., B. H. Cochran, C. D. Stiles, and P. Leder. 1983. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell 35:603-610. [DOI] [PubMed] [Google Scholar]
- 30.Kleinberger, T., and T. Shenk. 1993. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J. Virol. 67:7556-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremmer, E., K. Ohst, J. Kiefer, N. Brewis, and G. Walter. 1997. Separation of PP2A core enzyme and holoenzyme with monoclonal antibodies against the regulatory A subunit: abundant expression of both forms in cells. Mol. Cell. Biol. 17:1692-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lechward, K., S. Zolnierowicz, and B. A. Hemmings. 1999. Eukaryotic translation termination factor 1 associates with protein phosphatase 2A and targets it to ribosomes. Biochemistry (Moscow) 64:1373-1381. [PubMed] [Google Scholar]
- 33.Li, X., and D. M. Virshup. 2002. Two conserved domains in regulatory B subunits mediate binding to the A subunit of protein phosphatase 2A. Eur. J. Biochem. 269:546-552. [DOI] [PubMed] [Google Scholar]
- 34.Li, X., H. J. Yost, D. M. Virshup, and J. M. Seeling. 2001. Protein phosphatase 2A and its B56 regulatory subunit inhibit Wnt signaling in Xenopus. EMBO J. 20:4122-4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutterbach, B., and S. R. Hann. 1994. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol. Cell. Biol. 14:5510-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martens, E., I. Stevens, V. Janssens, J. Vermeesch, J. Gotz, J. Goris, and C. Van Hoof. 2004. Genomic organisation, chromosomal localisation tissue distribution and developmental regulation of the PR61/B′ regulatory subunits of protein phosphatase 2A in mice. J. Mol. Biol. 336:971-986. [DOI] [PubMed] [Google Scholar]
- 37.Mayer-Jaekel, R. E., H. Ohkura, P. Ferrigno, N. Andjelkovic, K. Shiomi, T. Uemura, D. M. Glover, and B. A. Hemmings. 1994. Drosophila mutants in the 55 kDa regulatory subunit of protein phosphatase 2A show strongly reduced ability to dephosphorylate substrates of p34cdc2. J. Cell Sci. 107:2609-2616. [DOI] [PubMed] [Google Scholar]
- 38.McCright, B., A. R. Brothman, and D. M. Virshup. 1996. Assignment of human protein phosphatase 2A regulatory subunit genes b56alpha, b56beta, b56gamma, b56delta, and b56epsilon (PPP2R5A-PPP2R5E), highly expressed in muscle and brain, to chromosome regions 1q41, 11q12, 3p21, 6p21.1, and 7p11.2→p12. Genomics 36:168-170. [DOI] [PubMed] [Google Scholar]
- 39.McCright, B., A. M. Rivers, S. Audlin, and D. M. Virshup. 1996. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J. Biol. Chem. 271:22081-22089. [DOI] [PubMed] [Google Scholar]
- 40.McCright, B., and D. M. Virshup. 1995. Identification of a new family of protein phosphatase 2A regulatory subunits. J. Biol. Chem. 270:26123-26128. [DOI] [PubMed] [Google Scholar]
- 41.Mumby, M. 1995. Regulation by tumour antigens defines a role for PP2A in signal transduction. Semin. Cancer Biol. 6:229-237. [DOI] [PubMed] [Google Scholar]
- 42.Nowak, S. J., C. Y. Pai, and V. G. Corces. 2003. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol. Cell. Biol. 23:6129-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamoto, K., C. Kamibayashi, M. Serrano, C. Prives, M. C. Mumby, and D. Beach. 1996. p53-dependent association between cyclin G and the B′ subunit of protein phosphatase 2A. Mol. Cell. Biol. 16:6593-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okamoto, K., H. Li, M. R. Jensen, T. Zhang, Y. Taya, S. S. Thorgeirsson, and C. Prives. 2002. Cyclin G recruits PP2A to dephosphorylate Mdm2. Mol. Cell 9:761-771. [DOI] [PubMed] [Google Scholar]
- 45.Pallas, D. C., L. K. Shahrik, B. L. Martin, S. Jaspers, T. B. Miller, D. L. Brautigan, and T. M. Roberts. 1990. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell 60:167-176. [DOI] [PubMed] [Google Scholar]
- 46.Patturajan, M., S. Nomoto, M. Sommer, A. Fomenkov, K. Hibi, R. Zangen, N. Poliak, J. Califano, B. Trink, E. Ratovitski, and D. Sidransky. 2002. DeltaNp63 induces beta-catenin nuclear accumulation and signaling. Cancer Cell 1:369-379. [DOI] [PubMed] [Google Scholar]
- 47.Pelengaris, S., T. Littlewood, M. Khan, G. Elia, and G. Evan. 1999. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell 3:565-577. [DOI] [PubMed] [Google Scholar]
- 48.Pulverer, B. J., C. Fisher, K. Vousden, T. Littlewood, G. Evan, and J. R. Woodgett. 1994. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene 9:59-70. [PubMed] [Google Scholar]
- 49.Ruediger, R., M. Hentz, J. Fait, M. Mumby, and G. Walter. 1994. Molecular model of the A subunit of protein phosphatase 2A: interaction with other subunits and tumor antigens. J. Virol. 68:123-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruediger, R., H. T. Pham, and G. Walter. 2001. Alterations in protein phosphatase 2A subunit interaction in human carcinomas of the lung and colon with mutations in the A beta subunit gene. Oncogene 20:1892-1899. [DOI] [PubMed] [Google Scholar]
- 51.Ruediger, R., H. T. Pham, and G. Walter. 2001. Disruption of protein phosphatase 2A subunit interaction in human cancers with mutations in the A alpha subunit gene. Oncogene 20:10-15. [DOI] [PubMed] [Google Scholar]
- 52.Ruediger, R., D. Roeckel, J. Fait, A. Bergqvist, G. Magnusson, and G. Walter. 1992. Identification of binding sites on the regulatory A subunit of protein phosphatase 2A for the catalytic C subunit and for tumor antigens of simian virus 40 and polyomavirus. Mol. Cell. Biol. 12:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rundell, K., and R. Parakati. 2001. The role of the SV40 ST antigen in cell growth promotion and transformation. Semin. Cancer Biol. 11:5-13. [DOI] [PubMed] [Google Scholar]
- 54.Ruvolo, P. P., W. Clark, M. Mumby, F. Gao, and W. S. May. 2002. A functional role for the B56 alpha-subunit of protein phosphatase 2A in ceramide-mediated regulation of Bcl2 phosphorylation status and function. J. Biol. Chem. 277:22847-22852. [DOI] [PubMed] [Google Scholar]
- 55.Schonthal, A. H. 2001. Role of serine/threonine protein phosphatase 2A in cancer. Cancer Lett 170:1-13. [DOI] [PubMed] [Google Scholar]
- 56.Sears, R., G. Leone, J. DeGregori, and J. R. Nevins. 1999. Ras enhances Myc protein stability. Mol. Cell 3:169-179. [DOI] [PubMed] [Google Scholar]
- 57.Sears, R., F. Nuckolls, E. Haura, Y. Taya, K. Tamai, and J. R. Nevins. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14:2501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sears, R., K. Ohtani, and J. R. Nevins. 1997. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol. Cell. Biol. 17:5227-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seeling, J. M., J. R. Miller, R. Gil, R. T. Moon, R. White, and D. M. Virshup. 1999. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science 283:2089-2091. [DOI] [PubMed] [Google Scholar]
- 60.Shtrichman, R., R. Sharf, H. Barr, T. Dobner, and T. Kleinberger. 1999. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc. Natl. Acad. Sci. USA 96:10080-10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shtrichman, R., R. Sharf, and T. Kleinberger. 2000. Adenovirus E4orf4 protein interacts with both Balpha and B′ subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Balpha. Oncogene 19:3757-3765. [DOI] [PubMed] [Google Scholar]
- 62.Sontag, E., S. Fedorov, C. Kamibayashi, D. Robbins, M. Cobb, and M. Mumby. 1993. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the MAP kinase pathway and induces cell proliferation. Cell 75:887-897. [DOI] [PubMed] [Google Scholar]
- 63.Strack, S., R. Ruediger, G. Walter, R. K. Dagda, C. A. Barwacz, and J. T. Cribbs. 2002. Protein phosphatase 2A holoenzyme assembly: identification of contacts between B-family regulatory and scaffolding A subunits. J. Biol. Chem. 277:20750-20755. [DOI] [PubMed] [Google Scholar]
- 64.Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy, G. R. Martin, and J. M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414:768-773. [DOI] [PubMed] [Google Scholar]
- 65.Turowski, P., B. Favre, K. S. Campbell, N. J. Lamb, and B. A. Hemmings. 1997. Modulation of the enzymatic properties of protein phosphatase 2A catalytic subunit by the recombinant 65-kDa regulatory subunit PR65alpha. Eur. J. Biochem. 248:200-208. [DOI] [PubMed] [Google Scholar]
- 66.Turowski, P., T. Myles, B. A. Hemmings, A. Fernandez, and N. J. Lamb. 1999. Vimentin dephosphorylation by protein phosphatase 2A is modulated by the targeting subunit B55. Mol. Biol. Cell. 10:1997-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Hoof, C., and J. Goris. 2004. PP2A fulfills its promises as tumor suppressor: which subunits are important? Cancer Cell 5:105-106. [DOI] [PubMed] [Google Scholar]
- 68.Voorhoeve, P. M., E. M. Hijmans, and R. Bernards. 1999. Functional interaction between a novel protein phosphatase 2A regulatory subunit, PR59, and the retinoblastoma-related p107 protein. Oncogene 18:515-524. [DOI] [PubMed] [Google Scholar]
- 69.Wadzinski, B. E., B. J. Eisfelder, L. F. Peruski, Jr., M. C. Mumby, and G. L. Johnson. 1992. NH2-terminal modification of the phosphatase 2A catalytic subunit allows functional expression in mammalian cells. J. Biol. Chem. 267:16883-16888. [PubMed] [Google Scholar]
- 70.Wadzinski, B. E., W. H. Wheat, S. Jaspers, L. F. Peruski, Jr., R. L. Lickteig, G. L. Johnson, and D. J. Klemm. 1993. Nuclear protein phosphatase 2A dephosphorylates protein kinase A-phosphorylated CREB and regulates CREB transcriptional stimulation. Mol. Cell. Biol. 13:2822-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, S. S., E. D. Esplin, J. L. Li, L. Huang, A. Gazdar, J. Minna, and G. A. Evans. 1998. Alterations of the PPP2R1B gene in human lung and colon cancer. Science 282:284-287. [DOI] [PubMed] [Google Scholar]
- 72.Welcker, M., A. Orian, J. Jin, J. A. Grim, J. W. Harper, R. N. Eisenman, and B. E. Clurman. 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA 101:9085-9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wera, S., A. Fernandez, N. J. Lamb, P. Turowski, M. Hemmings-Mieszczak, R. E. Mayer-Jaekel, and B. A. Hemmings. 1995. Deregulation of translational control of the 65-kDa regulatory subunit (PR65 alpha) of protein phosphatase 2A leads to multinucleated cells. J. Biol. Chem. 270:21374-21381. [DOI] [PubMed] [Google Scholar]
- 74.Yada, M., S. Hatakeyama, T. Kamura, M. Nishiyama, R. Tsunematsu, H. Imaki, N. Ishida, F. Okumura, K. Nakayama, and K. I. Nakayama. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan, Z., S. A. Fedorov, M. C. Mumby, and R. S. Williams. 2000. PR48, a novel regulatory subunit of protein phosphatase 2A, interacts with Cdc6 and modulates DNA replication in human cells. Mol. Cell. Biol. 20:1021-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, S. I., R. L. Lickteig, R. Estes, K. Rundell, G. Walter, and M. C. Mumby. 1991. Control of protein phosphatase 2A by simian virus 40 small-t antigen. Mol. Cell. Biol. 11:1988-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeh, E., M. Cunningham, H. Arnold, D. Chasse, T. Monteith, G. Ivaldi, W. C. Hahn, P. T. Stukenberg, S. Shenolikar, T. Uchida, C. M. Counter, J. R. Nevins, A. R. Means, and R. Sears. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6:308-318. [DOI] [PubMed] [Google Scholar]
- 78.Yu, J., A. Boyapati, and K. Rundell. 2001. Critical role for SV40 small-t antigen in human cell transformation. Virology 290:192-198. [DOI] [PubMed] [Google Scholar]
- 79.Zhao, Y., G. Boguslawski, R. S. Zitomer, and A. A. DePaoli-Roach. 1997. Saccharomyces cerevisiae homologs of mammalian B and B′ subunits of protein phosphatase 2A direct the enzyme to distinct cellular functions. J. Biol. Chem. 272:8256-8262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.