Figure 4.
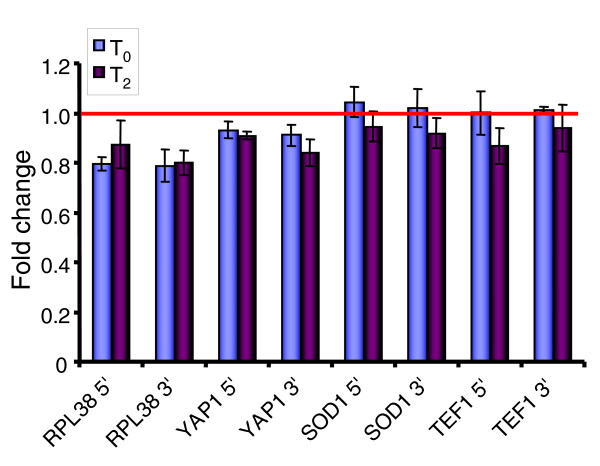
Quantitative RT-PCR analysis to detect presence of non-adenylated transcripts in T0 samples and samples 2 minutes after oxidative stress (T2). cDNA was synthesized using oligo-dT (to identify polyadenylated transcripts) or random hexamer primers. To determine if 5' or 3' ends of transcripts were more abundant, primer pairs were made to amplify 3' or 5' ends of each of four transcripts. Fold change represents the difference in abundance of 5' or 3' ends of transcripts in cDNAs synthesized using random hexamers versus oligo-dT primers. Measurements were obtained by quantitative RT-PCR and error bars represent the standard deviation of three measurements. The red horizontal bar at Fold Change = 1 indicates no difference in transcript abundance between oligo-dT-primed cDNA and random hexamer-primed cDNA. If non-adenylated transcripts were present, Fold Change > 1 would be expected.
