Figure 4.
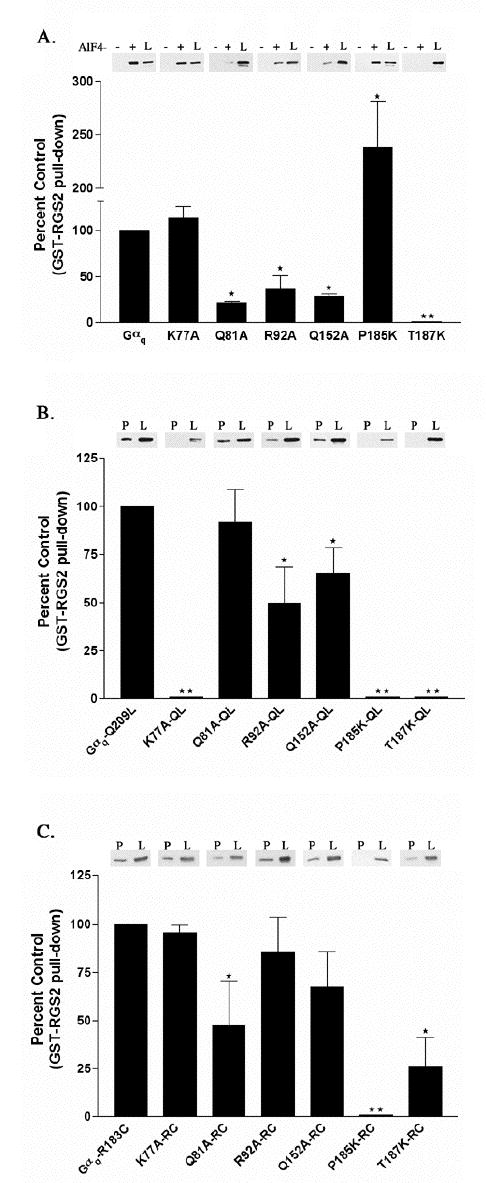
Interaction of GST-RGS2 with Gαq point mutants activated by AlF4−, the Q209L or the R183C mutation. (A) HEK-293 cells were transfected with EE tagged versions of Gαq point mutants and Gβ and Gγ constructs. Cells were lysed and binding to GST-RGS2 in the presence (+) or the absence (−) of AlF4− was determined as described in Experimental Procedures. The (+) and (−) lanes represent 40% of the Gαq or Gαq mutant pulled down from the 125 μl of lysate. In these experiments we detect little to no binding of GST- RGS2 to Gαq or Gαq point mutants in the absence of AlF4−. Underneath the representative western blot the percent of each Gαq mutant pulled down by GST- RGS2 in the presence of AlF4− is compared to the control, which is the percent of Gαq pulled down by GST-RGS2 in the presence of AlF4−, and is represented graphically as the percent of control ± S.D. (B) HEK-293 cells were transfected with EE tagged versions of Gαq-Q209L point mutants and Gβ and Gγ constructs. Cells were lysed and binding of the QL mutants to GST-RGS2 was determined as described in Experimental Procedures. Results are plotted as described in A. (C) HEK-293 cells were transfected with EE tagged versions of Gαq-R183C point mutants and Gβ and Gγ constructs. Cells were lysed and binding of the RC mutants to GST-RGS2 was determined as described in Experimental Procedures. The lanes labeled “P” in B and C represent 40% of the Gαq or Gαq mutant that was present in the pull-down from 200 μl of lysate. The lanes in A, B and C labeled “L” represent 4% of total Gαq or Gαq mutant available in the lysate for pull-down. Results are plotted as described in A. The (⋆) indicates that the amount of the marked Gαq mutant pulled-down is significantly different (p < 0.05) by one-way ANOVA followed by a Dunnett post-test, than the amount of Gαq pulled-down by GST-RGS2. The (⋆⋆) indicates that statistical analysis could not be performed on the binding of GST-RGS2 to the K77A-QL, P185K-QL, P185K-RC, T187K or T187K-QL mutants because there was no detectable pull-down. The data are averages from three to six independent experiments.
