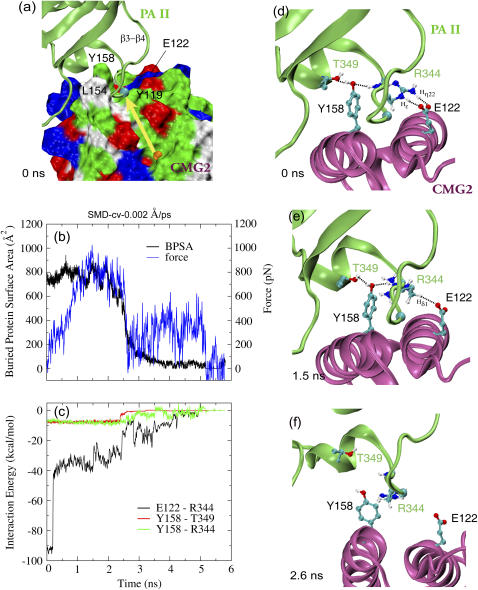
FIGURE 4.
Probing the binding site between PAII and CMG2 of the equilibrated neutral complex in a representative 0.002 Å/ps cv-SMD simulation. (a) Force is applied to the β-strands of PAII along the vector connecting the MIDAS ion (orange sphere) and the Cα atom (cyan sphere) of PAII Gly-342. The force direction is represented by a yellow arrow. CMG2 is shown in surface representation. The color code for positively charged, negatively charged, polar, and hydrophobic amino acids on the surface is blue, red, green, and white, respectively. (b) Profiles of applied force and resulting BPSA between PAII and CMG2 from the SMD simulation. (c) Interaction energy versus time profiles for three pairs of residues involved in binding between PAII and CMG2. (d–f) Snapshots of the PAII-CMG2 binding interface showing the disruption of the Arg-344PA-Glu-122CMG2 salt bridge and a couple of polar interactions between Tyr-158CMG2 and PAII.