Abstract
A primitive example of adaptation in gene expression is the balance between the rate of synthesis and degradation of cellular RNA, which allows rapid responses to environmental signals. Here, we investigate how multidrug efflux pump systems mediate the dynamics of a simple drug-inducible system in response to a steady level of inducer. Using fluorescence correlation spectroscopy, we measured in real time within a single bacterium the transcription activity at the RNA level of the acrAB-TolC multidrug efflux pump system. When cells are exposed to constant level of anhydrotetracycline inducer and are adsorbed onto a poly-L-lysine-coated surface, we found that the acrAB-TolC promoter is steadily active. We also monitored the activity of the tet promoter to characterize the effect of this efflux system on the dynamics of drug-inducible transcription. We found that the transcriptional response of the tet promoter to a steady level of aTc rises and then falls back to its preinduction level. The rate of RNA degradation was constant throughout the transcriptional pulse, indicating that the modulation of intracellular inducer concentration alone can produce this pulsating response. Single-cell experiments together with numerical simulations suggest that such pulsating response in drug-inducible genetic systems is a property emerging from the dependence of drug-inducible transcription on multidrug efflux systems.
INTRODUCTION
Single-celled, like multicelled, organisms exhibit properties essential for survival such as communication, homeostasis, and adaptation. A classic example of adaptation is the ability of bacteria to express nonspecific efflux pumps in response to antibiotic exposure. Efflux mechanisms are believed to be key determinants of antibiotic resistance (1,2). As a model system, we investigate the effect of a known efflux pump, the AcrAB-TolC system, on the dynamics of a simple tet-inducible promoter. Because the tetracycline is a known substrate of the AcrAB-TolC efflux pump (3), it is expected that the dynamics of tetracycline-inducible systems should depend strongly on this efflux pump. In this work, we characterize how the dynamics of the inducible tet promoter is associated with the promoter activity of the acrAB-TolC multidrug efflux pump system (4,5) in individual living bacteria of Escherichia coli.
Monitoring transcriptional responses in an isolated bacterium immobilized onto a surface has remained a technical challenge because they are often inaccessible to ensemble measurements (3,6). Most noninvasive characterizations of transcriptional activity in single cells use reporter proteins such as green fluorescent protein (GFP) or luciferase (7,8). However, the slow maturation time of GFP and the low light level of luciferase may not be well suited for the analysis of rapid transcriptional responses to an environmental stimulus. In a recent study (9), we proposed a noninvasive experimental approach to monitor the activity directly at the RNA level of any promoter in prokaryotes. In our preliminary study (9), we found that the activity of the tet promoter upon the steady level of induction exhibited an unexpected pulsating profile. Several questions thereafter arose: Are the pulses of activity caused by a variation of RNA degradation? What is the activity of the acrAB promoter during the induction process? Could we reproduce theoretically this pulsating behavior using simple hypotheses? To address these questions, we decided to develop a simple assay where the bacteria were attached on a poly-L-lysine. Under these conditions, cells cannot grow because they are tightly adsorbed to the coverslip, but they are still able to sense and respond to chemical inducers. These conditions are different from our preliminary study where cells were allowed to grow and divide on coverslips coated with agarose padding. We used the same apparatus as in Le et al. (9) to measure separately the activity of the promoter of the acrAB efflux system and of the tet promoter when cells are exposed to a steady level of anhydrotetracycline (aTc).
We used a dual plasmid system that was developed in Le et al. (9) to monitor in real time the transcription activity of the acrAB promoter in cell adsorbed onto a surface. A synthetic gene coding for a tandem of ms2-RNA binding sites was placed under the control of a chromosomal copy of the acrAB promoter (10) on one plasmid, while another plasmid was used to preexpress the MS2 coat protein fused to GFP (9,11). The ms2-RNA sequence has two identical specific binding sites for MS2-GFP protein. We preexpressed MS2-GFP by inducing with isopropyl B-D-thiogalactoside overnight. When the acrAB promoter is active, transcripts with two ms2-RNA binding sites are synthesized and the preexpressed MS2-GFP proteins bind to them. Because the ms2-RNA transcripts are fused to a ribosome binding site, their interaction with the ribosome makes the RNA/MS2-GFP complex diffuse 30-fold slower than the free GFP fusion protein (9). The relative concentrations of RNA/MS2-GFP complexes diffusing slowly and of free MS2-GFP proteins diffusing fast are measured using fluorescent correlation spectroscopy (FCS) (Supplementary Fig. 1) (9,12–14). This procedure is used to monitor the concentration of ms2-RNA transcripts placed under the control of the acrAB or tet promoters.
MATERIALS AND METHODS
Plasmids
Vectors based on the pZ family (15): 1), pZS12MS2-GFP (SC101 origin, 6–8 copies/cell, ampicillin, PLlacO-1 promoter)/pZE31ms2 (ColE1 origin, 50–70 copies/cell, ChlR, PLtetO-1 promoter); and 2), pZS12MS2-GFP (SC101 origin, 6–8 copies/cell, AmpR, PA1lacO-1 promoter)/pZE3acrABms2 in which the PLtetO-1 promoter from pZE31ms2 was replaced with the chromosomal acrAB promoter.
Cell strains
DH5αPRO (Clontech, Mountain View, CA): deoR, endA1, gyrA96, hsdR17(rk− mk+), recA1, relA1, supE44, thi-1, Δ(lacZYA-argF)U169, Φ80δlacZΔM15, F−, λ−, PN25/tetR, PlacIq/lacI, SpR.
Frag1A: F-, rha-, thi, gal, lacZam, ΔacrAB∷kanR, PN25/tetR, PlacIq/lacI, SpR.
Frag1B: F-, rha-, thi, gal, lacZam, PN25/tetR, Placiq/lacI, SpR.
The PN25/tetR, PlacIq/lacI, SpR cassette was transferred from DH5αPRO to Frag1 to generate Frag1B. The ΔacrAB:kanR cassette was transferred from KZM120 to Frag1B to generate Frag1A.
Growth condition
Cells carrying both reporter and expression plasmids were grown overnight at 30°C in M9 minimal salts (Qbiogene, Irvine, CA) supplemented with 0.1 mM CaCl2 + 2 mM MgSO4 + 0.4% glycerol + 0.5% casamino acids + 100 μg/ml Ampicillin + 34 μg/ml chloramphenicol + 50 μg/ml spectinomycin + 1 mM IPTG. Cells from overnight cultures were washed, diluted 20-fold, and regrown in fresh M9 media for an additional 2 h. The homogeneity and the level of cellular MS2-GFP expression were checked with fluorescence microscopy.
Determination of RNA concentration with FCS
We use the same setup and procedure described in Le et al. (9) and in the Supplementary Material. One MS2-GFP molecule in the detection volume within a living bacterium represents a concentration of 37 nM.
Transcription assay within a single cell
Cells were immobilized on a polylysine-coated coverslip in a reaction chamber. The chamber was filled with 200 μl of M9 media and placed on a thermocontrolled microscope stage set at 30°C. We used an FG loop deletion mutant of the coat protein of phage MS2 fused to GFP (denoted MS2-GFP), which binds a specific 23 nucleotides hairpin loop (MS2 binding site). In our experiments, MS2-GFP is preexpressed from an inducible promoter controlled by LacI. Cells were scanned using FCS for cells that were preexpressed MS2-GFP at the level of ∼11 μM. (MS2-GFP protein concentration is given for a homodimer, which is the MS2-RNA binding unit. Two MS2-GFP homodimers bind to one MS2-RNA transcript.) We replaced M9 media in the chamber with 200 μl of fresh M9 media premixed with inducer. FCS data were collected from cells at 5 min intervals for the first 25 min and 10 min intervals afterwards.
RNA decay in a single cell
Cells carrying the dual plasmids, pZS12-MS2GFP/pZE31-ms2, were induced to express ms2-RNA with 1 μg/ml aTc in the reaction chamber. At desired time intervals after induction, transcription was stopped with rifampicin. Rifampicin, premixed in M9 medium to a final concentration of 500 μg/ml, was used to rinse the reaction chamber three times to remove all residual aTc molecules. Finally, 200 μl of 500 μg/ml rifampicin in M9 was added to the reaction chamber to block transcription. RNA concentration was then measured with FCS at 2-min intervals.
Numerical simulations
Model assumptions include the following: 1), The equilibrium of the intracellular concentration of aTc with the external concentration after induction is instantaneous. For simplicity, we hypothesize that immediately after induction the effective efflux of aTc is constant with rate μ. 2), After induction, we assume that transcription and aTc efflux occur on longer timescales than repression and induction of the tet promoter. Accordingly, we use quasiequilibrium approximations for repression and induction kinetics (Supplementary Material).
RESULTS AND DISCUSSION
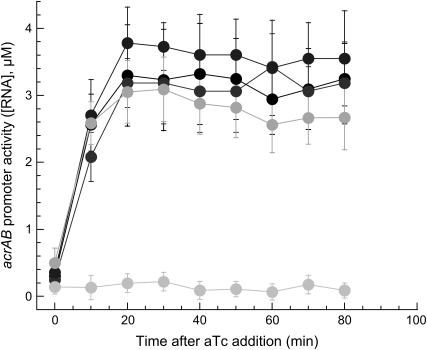
Since tetracycline is a known inducer for efflux systems (10,16), we first characterized the promoter activity of the acrAB promoter when Frag1B cells, wild-type for efflux system, were exposed to a steady level of the antibiotic inducer aTc. We monitored the activity of the acrAB promoter when E. coli cells were tightly adsorbed on a microscope slide coated with poly-L-lysine immersed in a growth medium (M9 minimal medium). Such irreversible adhesion of an individual bacterium onto a surface can induce major physiological responses (17), and under our conditions the cells did not grow or divide. We used glass coverslips coated with poly-L-lysine to promote a tight cellular attachment. The nonspecific absorption of bacteria on a poly-L-lysine-coated glass surface is mediated by electrostatic attraction between the bacterium and the coated surface (18). Although the precise details of the underlying interactions promoting cellular absorption are still subjects of research, poly-L-lysine is a bio-agent widely used for promoting cellular adhesion on hard substrates (19–21). Immediately after exposure to aTc, the activity of the acrAB promoter rose sharply for ∼10 min and leveled off to a steady plateau (Fig. 1). A series of studies showed that when E. coli cells are exposed to antibiotics, a multi-antibiotic resistance (mar) operon is upregulated. A gene product of this operon, marA, regulates the expression of more than 60 genes (22). In particular, it simultaneously activates its own transcription and that of the acrAB promoter, which constitutes a positive feedback loop (23). Therefore, exposure to aTc is expected to fully induce the expression of marA and acrAB because mar expression is self-reinforcing (3). The induction is self-reinforcing only if the aTc concentration has crossed a threshold concentration. The dynamics of this switch-like behavior may also be limited by the fact that marA needs to be synthesized to activate the acrAB promoter, which may contribute to lengthen the timescale of acrAB activation down to the observed 10 min.
FIGURE 1.
Real-time acrAB promoter activity (RNA concentration) in four wild-type cells (Frag1B) induced with 400 ng/ml aTc and one cell without aTc (flat trace). Cells were immobilized on polylysine-coated surfaces. Error bars represent uncertainties in the fit parameters extracted from the autocorrelation function (Methods).
To characterize the effect of the AcrAB efflux system on drug-inducible gene expression, we monitored the activity of the tet promoter when a bacterium was exposed to a steady level of antibiotic (aTc). As described previously, cells were immobilized on a surface coated with poly-L-lysine and couldn't grow or divide but exhibited transcriptional responses to various levels of inducer for several hours after being adsorbed to the coated glass surface (24). We transformed E. coli strain DH5αPRO with two plasmids. One plasmid was used to express a MS2 phage coat protein fused to GFP (11,25,26) (MS2-GFP) from a LacI-controlled promoter. The other plasmid was used to express the ms2-RNA sequence from a TetR-controlled promoter (15). After induction with aTc, the concentration of ms2-RNA transcripts rose sharply and peaked at ∼20 min then fell back to the preinduction level. We then characterized the response of this system when cells were exposed to various levels of the inducer aTc (Fig. 2, A–C). Since the ms2-RNA level for a given inducer concentration varied from cell to cell, we plotted the mean of the responses from 10 single cells to also show that the average transcriptional response increases with the level of inducer. When cells were exposed to different levels of inducer (200, 400, 1000 ng/ml), the initial rate of ms2-RNA synthesis measured from the population was similar for all aTc concentrations, suggesting that the initial rate of ms2-RNA synthesis was maximal (Fig. 2 D).
FIGURE 2.
Real-time tet promoter activity from 10 single cells induced at (A), 200 ng/ml aTc, (B), 400 ng/ml aTc, and (C), 1 μg/ml aTc. Error bars represent uncertainties in the fit parameters extracted from the autocorrelation function. (D) Average profiles of ms2-RNA concentration from 10 single cells. No induction (light gray), 200 ng/ml aTc (gray), 400 ng/ml aTc (dark gray), and 1 μg/ml aTc (black). Cells were immobilized on polylysine-coated surfaces. Error bars represent the standard deviations from the ms2-RNA concentration distributions across a population of 10 cells at a given time point.
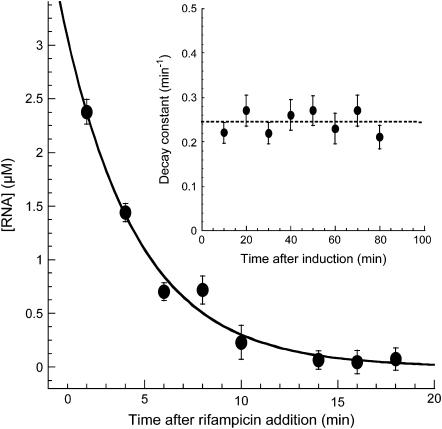
The observed pulsating transcriptional response could be produced by the temporal variations of either one or both: synthesis and degradation of RNA. To analyze the contribution of ms2-RNA degradation to the observed transcriptional response, we blocked transcription with addition of rifampicin. The antibiotic rifampicin irreversibly blocks transcription of ms2-RNA (27). Subsequently, we measured the degradation rate of ms2-RNA transcripts in single cells at various time points after addition of rifampicin (Fig. 3). We found that RNA degradation could be approximated by a simple exponential decay and was independent of both the time intervals after induction and RNA abundance (data not shown) (28) (Fig. 3, inset). This result suggests that modulation of RNA synthesis, not RNA degradation, contributes to the observed pulsating response to aTc exposure.
FIGURE 3.
RNA decay in individual E. coli cells. DH5αPRO cells induced with 1 μg/ml aTc. Transcription was blocked with the addition of rifampicin at various time points after induction. Decay of RNA concentration in a single cell exposed to 500 μg/ml of rifampicin at 30 min after induction (•). Exponential fit with a decay constant of 0.22 min−1 (solid line). Error bars represent uncertainties in the fit parameters of the associated autocorrelation functions (Methods). (Inset) RNA degradation rate as a function of time from eight cells (dashed line; average degradation rate, 0.26 min−1). Error bars represent fitting uncertainties in the decay constant.
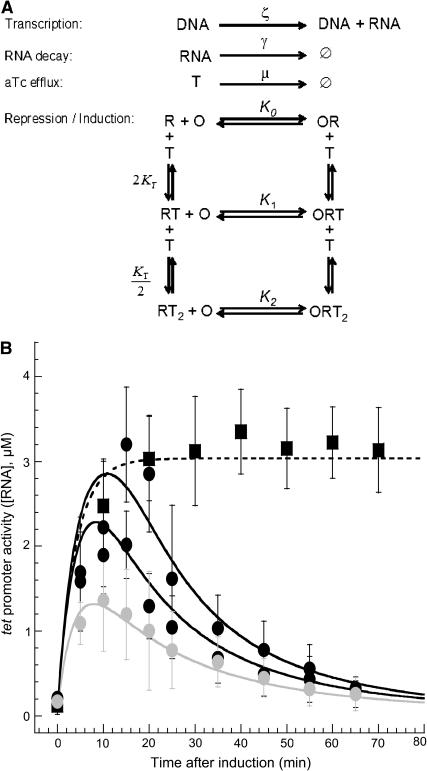
Next, we hypothesize that the observed transcriptional pulses are caused by the modulation of TetR's repression activity (15,29). Although the extracellular concentration of aTc inducer is constant throughout the measurements, the intracellular concentration of aTc may vary as a function of time (3,6,30). To gain further insight into the observed profile of induced RNA, we simulated the dynamics of transcription, repression, and RNA degradation in an individual living cell that did not grow or divide (Fig. 4 A). In this simple model, we assumed that the modulation of RNA synthesis was caused solely by the temporal variation of intracellular concentration of inducer. In these simulations, we assumed for simplicity that the intracellular concentration was equal to the extracellular concentration of inducer at the initial time point and after this time the intracellular concentration of inducer decreases with a constant rate. Using published values (Supplementary Material) of kinetic parameters for the aTc-tet system, plasmid copy number, and our measured RNA degradation rate (Fig. 3), the transcriptional pulse in response to antibiotic exposure was qualitatively recovered. In contrast, when the efflux rate was set to zero, the intracellular concentration of inducer remained equal to the extracellular concentration of inducer and the associated induced RNA concentration reached a steady plateau (Fig. 4 B).
FIGURE 4.
Model of inducible transcription in a living cell immobilized on a polylysine-coated surface. (A) Simple reaction scheme of efflux-mediated inducible transcription. T, R, and O represent aTc molecules, TetR dimers, and operator sites, respectively. The tet promoter has two identical operators. RT, ORT, RT2, and ORT2 stand for TetR:aTc, O:TetR:aTc, TetR:aTc2, and O:TetR:aTc2 complexes, respectively. K0, K1, K2, and KT are binding constants (Supplementary Material). (B) Simulated RNA profiles (solid lines) in wild-type cells for various aTc induction levels: 200 ng/ml (light shaded), 400 ng/ml (shaded), and 1000 ng/ml (dark shaded). Experimental data from Fig. 2 D (•). Simulated RNA profile (dashed line) for steady intracellular [aTc] = 400 ng/ml (μ = 0 s−1). Experimental data from Fig. 5 A (▪).
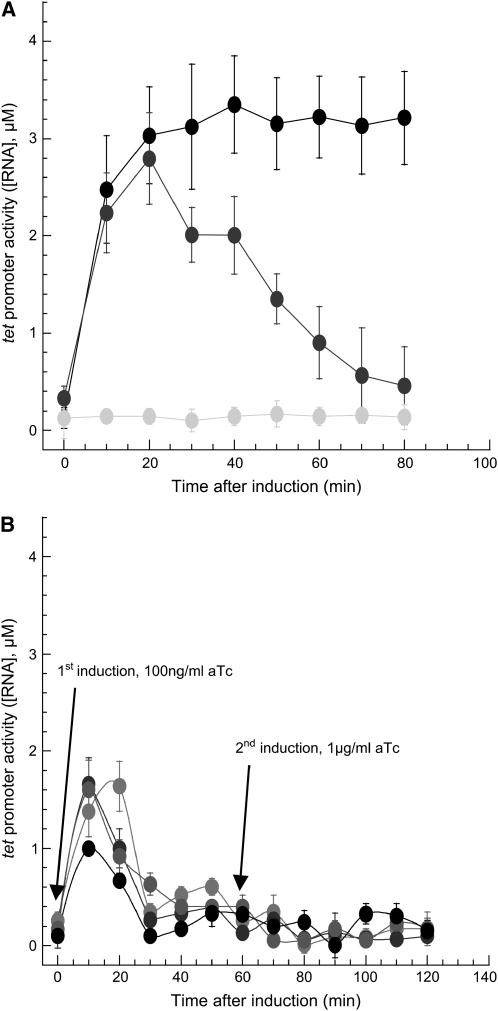
We recently showed that the deletion of acrAB in dividing cells altered the pulsating transcriptional dynamics of a tet-inducible gene and caused overexpression of induced RNA (9). Consequently, we also found that the expression level of proteins controlled by a tet-inducible promoter is severalfold higher in ΔacrAB efflux mutant than in wild-type cells. Here, we performed similar transcription assays in the ΔacrAB mutant cells (16), Frag1A (Methods). In this mutant, the AcrAB-TolC multidrug efflux pump system, which is a key factor for multidrug resistance and thus controls the intracellular concentration of inducer (3,6), was defective. This mutant strain was transformed with the same dual plasmid system as in Le et al. (9), and we monitored the transcription of the ms2-RNA gene from a tet promoter as a function of time. We found that after induction with aTc (400 ng/ml), the concentration of induced ms2-RNA transcripts from Frag1A cells exceeded the one obtained from Frag1B cells, wild-type for the efflux system (Fig. 5 A). The fact that RNA concentration reaches a steady plateau in mutant cells indicates that the intracellular aTc remains present in the cytoplasm and might not be as rapidly expelled by the efflux pump as in wild-type cells. Our model confirms this hypothesis: the response of mutant cells can be fitted by simply setting the efflux rate to zero while keeping all other parameters unchanged (Fig. 4 B). This observation suggests that the acrAB-TolC multidrug efflux system contributes to the modulation of the intracellular concentration of inducer, which consequently regulates the transcription activity of inducible genetic systems to produce an adaptive response (32,33). In particular, we saw that when cells are tightly adsorbed onto a surface and exposed to the inducer, the acrAB promoter remains active and is possibly responsible for the efflux of the antibiotic aTc. To check this hypothesis, after an initial induction with aTc (100 ng/ml), we exposed the cells to a second, much larger level of induction (1000 ng/ml). Under this condition, the cells did not respond to the second induction with aTc (Fig. 5 B). There may be several, combined reasons responsible for the observed result. The first hypothesis is that the number of pumps accumulates in the cell after the first induction. But the effective efflux rate needs to be several orders of magnitude higher than the one initially measured during the first induction to pump out the inducer faster than the time it takes to form transcripts. The second hypothesis is to assume that some influx mechanism importing the inducer remains inactive after the first induction. The combination of both hypotheses could provide an efficient mechanism to drastically decrease the effective membrane permeability to antibiotics after the first exposure. It is also worth noting that upon exposure to antibiotics, marA activates the synthesis of the acrAB efflux pump but also induces the synthesis of the small RNA micF. The small RNA micF is a translational repressor of the ompF influx system, which is believed to be the principal pathway for β-lactam antibiotics pathway (34,35). The combined activation of acrAB synthesis and inhibition of ompF translation is also compatible with the result plotted in Fig. 5 B.
FIGURE 5.
Efflux-mediated inducible transcription. (A) Concentration profiles of ms2-RNA transcripts from an inducible tet promoter in a Frag1A mutant cell (ΔacrAB) without aTc (shaded) and with 400 ng/ml aTc (solid). Wild-type Frag 1B cell with 400 ng/ml (dark shaded). (B) Tet promoter activity with sequential inductions in four nondividing wild-type cells (Frag1B). Cells were first induced with 100 ng/ml aTc at time zero followed by a second induction with 1 μg/ml aTc at time 60 min after first induction. Arrows indicate times of induction. Error bars represent uncertainties in the fit parameters extracted from the autocorrelation function.
This study illustrates how cellular activities, such as efflux mechanisms, can shape the dynamics of inducible transcriptional circuits to produce an adaptive response (36). Such interplay among regulatory systems imposes specific constraints on the modeling of inducible transcriptional networks. It is conceivable that the dependence of one process on another constitutes a generic design principle of regulatory organization (33,37). In this picture, such regulatory organization may permit rapid adaptation to a changing environment without requiring long-lasting mutational processes. Our study suggests that the observed pulsating behavior in inducible transcription is itself a systems property arising from the dependence of gene expression on multidrug resistance determinants.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
We thank Wolfgang Hillen's laboratory (Lehrstuhl für Mikrobiologie, Institut für Biologie, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany) for help with aTc reaction rates and T. Pan and T. Griggs for helpful comments on the manuscript.
This work was partially supported by a National Institutes of Health training grant and by the Materials Research Science and Engineering Center and Institute for Biophysical Dynamics seed fund. T.E. acknowledges partial support by joint research funding under H.28 of the U.S. Dept. of Energy Contract W-31-109-ENG-38.
References
- 1.Neyfakh, A. A. 2002. Mystery of multidrug transporters: the answer can be simple. Mol. Microbiol. 44:1123–1130. [DOI] [PubMed] [Google Scholar]
- 2.Markham, P. N., and A. A. Neyfakh. 2001. Efflux-mediated drug resistance in Gram-positive bacteria. Curr. Opin. Microbiol. 4:509–514. [DOI] [PubMed] [Google Scholar]
- 3.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs. 64:159–204. [DOI] [PubMed] [Google Scholar]
- 4.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis, K. 2005. Persister cells and the riddle of biofilm survival. Biochemistry (Mosc.). 70:267–274. [DOI] [PubMed] [Google Scholar]
- 6.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science. 300:976–980. [DOI] [PubMed] [Google Scholar]
- 7.Bhaumik, S., and S. S. Gambhir. 2002. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc. Natl. Acad. Sci. USA. 99:377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalir, S., J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M. G. Surette, and U. Alon. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science. 292:2080–2083. [DOI] [PubMed] [Google Scholar]
- 9.Le, T. T., S. Harlepp, C. C. Guet, K. Dittmar, T. Emonet, T. Pan, and P. Cluzel. 2005. Real-time RNA profiling within a single bacterium. Proc. Natl. Acad. Sci. USA. 102:9160–9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101–112. [DOI] [PubMed] [Google Scholar]
- 11.Bertrand, E., P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer, and R. M. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 2:437–445. [DOI] [PubMed] [Google Scholar]
- 12.Cluzel, P., M. Surette, and S. Leibler. 2000. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 287:1652–1655. [DOI] [PubMed] [Google Scholar]
- 13.Eigen, M., and R. Rigler. 1994. Sorting single molecules—application to diagnostics and evolutionary biotechnology. Proc. Natl. Acad. Sci. USA. 91:5740–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauer, B., E. Neumann, J. Widengren, and R. Rigler. 1996. Fluorescence correlation spectrometry of the interaction kinetics of tetramethylrhodamin alpha-bungarotoxin with Torpedo californica acetylcholine receptor. Biophys. Chem. 58:3–12. [DOI] [PubMed] [Google Scholar]
- 15.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I-1-I-2 regulatory elements. Nucleic Acids Res. 25:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45–55. [DOI] [PubMed] [Google Scholar]
- 17.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA. 99:2287–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke, M., G. Schatten, D. Mazia, and J. A. Spudich. 1975. Visualization of actin fibers associated with the cell membrane in amoebae of Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 72:1758–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agladze, K., D. Jackson, and T. Romeo. 2003. Periodicity of cell attachment patterns during Escherichia coli biofilm development. J. Bacteriol. 185:5632–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowan, S. E., D. Liepmann, and J. D. Keasling. 2001. Development of engineered biofilms on poly-L-lysine patterned surfaces. Biotechnol. Lett. 23:1235–1241. [Google Scholar]
- 21.Gracia, E., A. Fernandez, P. Conchello, J. L. Alabart, M. Perez, and B. Amorena. 1999. In vitro development of Staphylococcus aureus biofilms using slime-producing variants and ATP-bioluminescence for automated bacterial quantification. Luminescence. 14:23–31. [DOI] [PubMed] [Google Scholar]
- 22.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowe, D. C. D., and D. K. Summers. 1999. The quiescent-cell expression system for protein synthesis in Escherichia coli. Appl. Environ. Microbiol. 65:2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beach, D. L., E. D. Salmon, and K. Bloom. 1999. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9:569–578. [DOI] [PubMed] [Google Scholar]
- 26.Peabody, D. S., and K. R. Ely. 1992. Control of translational repression by protein-protein interactions. Nucleic Acids Res. 20:1649–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA. 99:9697–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193–227. [DOI] [PubMed] [Google Scholar]
- 29.Lederer, T., M. Takahashi, and W. Hillen. 1995. Thermodynamic analysis of tetracycline-mediated induction of Tet repressor by a quantitative methylation protection assay. Anal. Biochem. 232:190–196. [DOI] [PubMed] [Google Scholar]
- 30.McMurry, L., R. E. Petrucci Jr., and S. B. Levy. 1980. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. USA. 77:3974–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reference deleted in proof.
- 32.Goh, E. B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA. 99:17025–17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirschner, M., and J. Gerhart. 1998. Evolvability. Proc. Natl. Acad. Sci. USA. 95:8420–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thanassi, D. G., G. S. Suh, and H. Nikaido. 1995. Role of outer membrane barrier in efflux-mediated tetracycline resistance of Escherichia coli. J. Bacteriol. 177:998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nestorovich, E. M., C. Danelon, M. Winterhalter, and S. M. Bezrukov. 2002. Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc. Natl. Acad. Sci. USA. 99:9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guet, C. C., M. B. Elowitz, W. Hsing, and S. Leibler. 2002. Combinatorial synthesis of genetic networks. Science. 296:1466–1470. [DOI] [PubMed] [Google Scholar]
- 37.Conrad, M. 1990. The geometry of evolution. Biosystems. 24:61–81. [DOI] [PubMed] [Google Scholar]